1L40
 
 | |
7M5L
 
 | |
7JG1
 
 | | Dimeric Immunoglobin A (dIgA) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Igh protein, ... | | 著者 | Kumar Bharathkar, S, Parker, B.P, Malyutin, A.G, Stadtmueller, B.M. | | 登録日 | 2020-07-18 | | 公開日 | 2020-11-11 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | The structures of Secretory and dimeric Immunoglobulin A.
Elife, 9, 2020
|
|
1L47
 
 | |
7M5N
 
 | | PCNA bound to peptide mimetic with linker | | 分子名称: | 1,3-dimethylbenzene, Peptide mimetic (ACE)RQCSMTCFYHSK(NH2) with linker, Proliferating cell nuclear antigen | | 著者 | Vandborg, B.A, Bruning, J.B. | | 登録日 | 2021-03-24 | | 公開日 | 2021-08-11 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.11 Å) | | 主引用文献 | A cell permeable bimane-constrained PCNA-interacting peptide.
Rsc Chem Biol, 2, 2021
|
|
5GL3
 
 | |
7MJ6
 
 | | HLA-A*02:01 bound to Neuroblastoma Derived IGFBPL1 peptide | | 分子名称: | Beta-2-microglobulin, GLYCEROL, Insulin-like growth factor-binding protein-like 1 peptide, ... | | 著者 | Toor, J.S, Tripathi, S.M, Truong, H.V, Yarmarkovich, M, Maris, J.M, Sgourakis, N.G. | | 登録日 | 2021-04-19 | | 公開日 | 2021-08-18 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Cross-HLA targeting of intracellular oncoproteins with peptide-centric CARs.
Nature, 599, 2021
|
|
5VYQ
 
 | | Crystal structure of the N-formyltransferase Rv3404c from mycobacterium tuberculosis in complex with YDP-Qui4N and folinic acid | | 分子名称: | 1,2-ETHANEDIOL, 3-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]PROPANE-1-SULFONIC ACID, CHLORIDE ION, ... | | 著者 | Dunsirn, M.M, Thoden, J.B, Holden, H.M. | | 登録日 | 2017-05-26 | | 公開日 | 2017-07-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Biochemical Investigation of Rv3404c from Mycobacterium tuberculosis.
Biochemistry, 56, 2017
|
|
1L50
 
 | |
6GQT
 
 | | KRAS-169 Q61H GPPNHP + PPIN-2 | | 分子名称: | CITRIC ACID, GTPase KRas, MAGNESIUM ION, ... | | 著者 | Cruz-Migoni, A, Canning, P, Quevedo, C.E, Carr, S.B, Phillips, S.E.V, Rabbitts, T.H. | | 登録日 | 2018-06-08 | | 公開日 | 2019-02-06 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Structure-based development of new RAS-effector inhibitors from a combination of active and inactive RAS-binding compounds.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5VV8
 
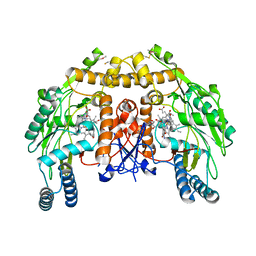 | | Structure of bovine endothelial nitric oxide synthase heme domain in complex with 4-(2-(((2-Aminoquinolin-7-yl)methyl)amino)ethyl)-2-methylbenzonitrile | | 分子名称: | 4-(2-{[(2-aminoquinolin-7-yl)methyl]amino}ethyl)-2-methylbenzonitrile, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | 著者 | Li, H, Poulos, T.L. | | 登録日 | 2017-05-19 | | 公開日 | 2017-08-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Hydrophilic, Potent, and Selective 7-Substituted 2-Aminoquinolines as Improved Human Neuronal Nitric Oxide Synthase Inhibitors.
J. Med. Chem., 60, 2017
|
|
1L38
 
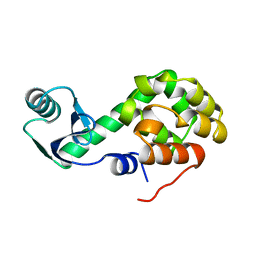 | |
4PV2
 
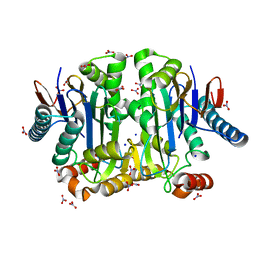 | | Crystal structure of potassium-dependent plant-type L-asparaginase from Phaseolus vulgaris in complex with K+ and Na+ cations | | 分子名称: | L-ASPARAGINASE ALPHA SUBUNIT, L-ASPARAGINASE BETA SUBUNIT, NITRATE ION, ... | | 著者 | Bejger, M, Gilski, M, Imiolczyk, B, Clavel, D, Jaskolski, M. | | 登録日 | 2014-03-14 | | 公開日 | 2014-09-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Na+/K+ exchange switches the catalytic apparatus of potassium-dependent plant L-asparaginase
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1L45
 
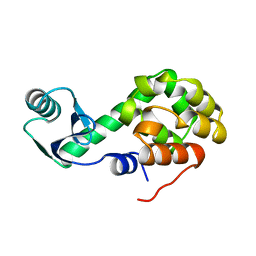 | |
5VVC
 
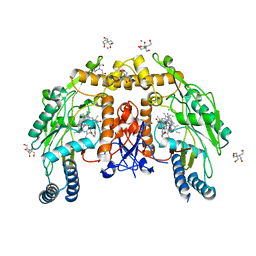 | | Structure of human endothelial nitric oxide synthase heme domain in complex with 4-(2-(((2-Amino-4-methylquinolin-7-yl)methyl)amino)ethyl)-2-methylbenzonitrile | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-{[(2-amino-4-methylquinolin-7-yl)methyl]amino}ethyl)-2-methylbenzonitrile, 5,6,7,8-TETRAHYDROBIOPTERIN, ... | | 著者 | Chreifi, G, Li, H, Poulos, T.L. | | 登録日 | 2017-05-19 | | 公開日 | 2017-08-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Hydrophilic, Potent, and Selective 7-Substituted 2-Aminoquinolines as Improved Human Neuronal Nitric Oxide Synthase Inhibitors.
J. Med. Chem., 60, 2017
|
|
1L69
 
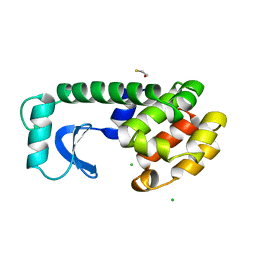 | |
4BY9
 
 | | The structure of the Box CD enzyme reveals regulation of rRNA methylation | | 分子名称: | 5'-R(*UP*CP*GP*CP*CP*CP*AP*UP*CP*AP*CP)-3', 50S RIBOSOMAL PROTEIN L7AE, FIBRILLARIN-LIKE RRNA/TRNA 2'-O-METHYLTRANSFERASE, ... | | 著者 | Lapinaite, A, Simon, B, Skjaerven, L, Rakwalska-Bange, M, Gabel, F, Carlomagno, T. | | 登録日 | 2013-07-18 | | 公開日 | 2013-10-09 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Structure of the Box C/D Enzyme Reveals Regulation of RNA Methylation.
Nature, 502, 2013
|
|
4JNE
 
 | | Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, Hsp70 CHAPERONE DnaK, ... | | 著者 | Qi, R, Sarbeng, E.B, Liu, Q, Le, K.Q, Xu, X, Xu, H, Yang, J, Wong, J.L, Vorvis, C, Hendrickson, W.A, Zhou, L, Liu, Q. | | 登録日 | 2013-03-15 | | 公開日 | 2013-05-29 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP.
Nat.Struct.Mol.Biol., 20, 2013
|
|
6KNK
 
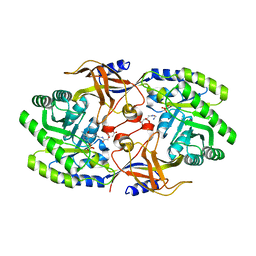 | | Crystal structure of SbnH in complex with citryl-diaminoethane | | 分子名称: | (2S)-2-{2-[(2-AMINOETHYL)AMINO]-2-OXOETHYL}-2-HYDROXYBUTANEDIOIC ACID, (2~{S})-2-[2-[2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]ethylamino]-2-oxidanylidene-ethyl]-2-oxidanyl-butanedioic acid, PHOSPHATE ION, ... | | 著者 | Tang, J, Ju, Y, Zhou, H. | | 登録日 | 2019-08-05 | | 公開日 | 2019-11-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural Insights into Substrate Recognition and Activity Regulation of the Key Decarboxylase SbnH in Staphyloferrin B Biosynthesis.
J.Mol.Biol., 431, 2019
|
|
4AMF
 
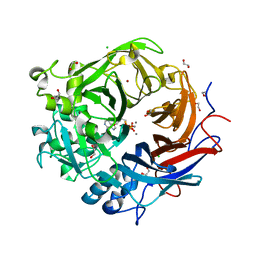 | | Pseudomonas fluorescens PhoX in complex with the substrate analogue AppCp | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Yong, S.C, Roversi, P, Lillington, J.E.D, Zeldin, O.B, Garman, E.F, Lea, S.M, Berks, B.C. | | 登録日 | 2012-03-09 | | 公開日 | 2013-03-20 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | A Complex Iron-Calcium Cofactor Catalyzing Phosphotransfer Chemistry
Science, 345, 2014
|
|
7FF8
 
 | |
4PVT
 
 | | Crystal Structure of VIM-2 metallo-beta-lactamase in complex with ML302F | | 分子名称: | (2Z)-2-sulfanyl-3-(2,3,6-trichlorophenyl)prop-2-enoic acid, Beta-lactamase class B VIM-2, FORMIC ACID, ... | | 著者 | Aik, W.S, Brem, J, McDonough, M.A, Schofield, C.J. | | 登録日 | 2014-03-18 | | 公開日 | 2014-11-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Rhodanine hydrolysis leads to potent thioenolate mediated metallo-beta-lactamase inhibition.
Nat.Chem., 6, 2014
|
|
2X0R
 
 | | R207S, R292S Mutant of Malate Dehydrogenase from the Halophilic Archeon Haloarcula marismortui (HoloForm) | | 分子名称: | CHLORIDE ION, MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Irimia, A, Ebel, C, Vellieux, F.M.D, Richard, S.B, Cosenza, L.W, Zaccai, G, Madern, D. | | 登録日 | 2009-12-17 | | 公開日 | 2009-12-29 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.915 Å) | | 主引用文献 | The Oligomeric States of Haloarcula Marismortui Malate Dehydrogenase are Modulated by Solvent Components as Shown by Crystallographic and Biochemical Studies
J.Mol.Biol., 326, 2003
|
|
6D09
 
 | | Crystal structure of PT2440 bound to HIF2a-B*:ARNT-B* complex | | 分子名称: | 3-{[(3R)-4-(difluoromethyl)-2,2-difluoro-3-hydroxy-1,1-dioxo-2,3-dihydro-1H-1-benzothiophen-5-yl]oxy}-5-fluorobenzonitrile, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | 著者 | Du, X. | | 登録日 | 2018-04-10 | | 公開日 | 2018-10-31 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal structure of PT1940 bound to HIF2a-B*:ARNT-B* complex
To Be Published
|
|
5ZBH
 
 | | The Crystal Structure of Human Neuropeptide Y Y1 Receptor with BMS-193885 | | 分子名称: | Neuropeptide Y receptor type 1,T4 Lysozyme,Neuropeptide Y receptor type 1, dimethyl 4-{3-[({3-[4-(3-methoxyphenyl)piperidin-1-yl]propyl}carbamoyl)amino]phenyl}-2,6-dimethyl-1,4-dihydropyridine-3,5-dicarboxylate | | 著者 | Yang, Z, Han, S, Zhao, Q, Wu, B. | | 登録日 | 2018-02-11 | | 公開日 | 2018-04-25 | | 最終更新日 | 2018-05-09 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural basis of ligand binding modes at the neuropeptide Y Y1receptor
Nature, 556, 2018
|
|
