6L1C
 
 | |
6L1I
 
 | |
6U97
 
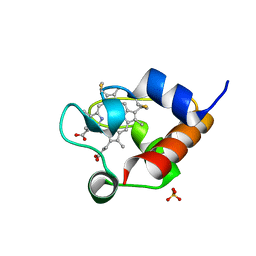 | | Structure of OmcF_H47I mutant | | 分子名称: | Lipoprotein cytochrome c, 1 heme-binding site, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Pokkuluri, P.R. | | 登録日 | 2019-09-06 | | 公開日 | 2020-02-19 | | 実験手法 | X-RAY DIFFRACTION (1.13 Å) | | 主引用文献 | Modulation of the Redox Potential and Electron/Proton Transfer Mechanisms in the Outer Membrane Cytochrome OmcF FromGeobacter sulfurreducens.
Front Microbiol, 10, 2019
|
|
6KZ7
 
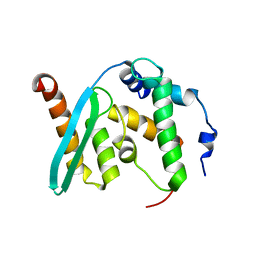 | | The crystal structure of BAF155 SWIRM domain and N-terminal elongated hSNF5 RPT1 domain complex: Chromatin remodeling complex | | 分子名称: | SWI/SNF complex subunit SMARCC1, SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1 | | 著者 | Lee, W, Han, J, Kim, I, Park, J.H, Joo, K, Lee, J, Suh, J.Y. | | 登録日 | 2019-09-23 | | 公開日 | 2020-07-08 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | A Coil-to-Helix Transition Serves as a Binding Motif for hSNF5 and BAF155 Interaction.
Int J Mol Sci, 21, 2020
|
|
5IH2
 
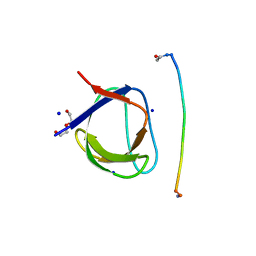 | | Structure, thermodynamics, and the role of conformational dynamics in the interactions between the N-terminal SH3 domain of CrkII and proline-rich motifs in cAbl | | 分子名称: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, Adapter molecule crk, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Bhatt, V.S, Zeng, D, Krieger, I, Sacchettini, J.C, Cho, J.-H. | | 登録日 | 2016-02-28 | | 公開日 | 2016-06-29 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Binding Mechanism of the N-Terminal SH3 Domain of CrkII and Proline-Rich Motifs in cAbl.
Biophys.J., 110, 2016
|
|
1JB6
 
 | |
6L10
 
 | | PHF20L1 Tudor1 - MES | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, PHD finger protein 20-like protein 1, SULFATE ION | | 著者 | Lv, M.Q, Gao, J. | | 登録日 | 2019-09-27 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
1C1Z
 
 | | CRYSTAL STRUCTURE OF HUMAN BETA-2-GLYCOPROTEIN-I (APOLIPOPROTEIN-H) | | 分子名称: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA2-GLYCOPROTEIN-I, ... | | 著者 | Schwarzenbacher, R, Zeth, K, Diederichs, K, Gries, A, Kostner, G.M, Laggner, P, Prassl, R. | | 登録日 | 1999-07-22 | | 公開日 | 1999-11-19 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.87 Å) | | 主引用文献 | Crystal structure of human beta2-glycoprotein I: implications for phospholipid binding and the antiphospholipid syndrome.
EMBO J., 18, 1999
|
|
6L1F
 
 | |
6UVJ
 
 | |
6UWX
 
 | |
6UVM
 
 | | Cocrystal of BRD4(D1) with a methyl carbamate thiazepane inhibitor | | 分子名称: | 1,2-ETHANEDIOL, 1-[(7S)-7-(thiophen-2-yl)-6,7-dihydro-1,4-thiazepin-4(5H)-yl]ethan-1-one, Bromodomain-containing protein 4 | | 著者 | Johnson, J.A, Pomerantz, W.C.K. | | 登録日 | 2019-11-03 | | 公開日 | 2020-01-01 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Evaluating the Advantages of Using 3D-Enriched Fragments for Targeting BET Bromodomains.
Acs Med.Chem.Lett., 10, 2019
|
|
1BJA
 
 | | ACTIVATION DOMAIN OF THE PHAGE T4 TRANSCRIPTION FACTOR MOTA | | 分子名称: | SULFATE ION, TRANSCRIPTION REGULATORY PROTEIN MOTA | | 著者 | Finnin, M.S, Cicero, M.P, Davies, C, Porter, S.J, White, S.W, Kreuzer, K.N. | | 登録日 | 1998-06-23 | | 公開日 | 1998-11-04 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | The activation domain of the MotA transcription factor from bacteriophage T4.
EMBO J., 16, 1997
|
|
6JCG
 
 | | Room temperature structure of HIV-1 Integrase catalytic core domain by serial femtosecond crystallography. | | 分子名称: | CACODYLATE ION, Integrase | | 著者 | Park, J.H, Shi, Y, Han, J, Li, X, Kim, T.H, Yun, J.H. | | 登録日 | 2019-01-28 | | 公開日 | 2019-07-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Non-Cryogenic Structure and Dynamics of HIV-1 Integrase Catalytic Core Domain by X-ray Free-Electron Lasers.
Int J Mol Sci, 20, 2019
|
|
6VMZ
 
 | | Crystal Structure of a H5N1 influenza virus hemagglutinin with CBS1117 | | 分子名称: | 2,6-dichloro-N-[1-(propan-2-yl)piperidin-4-yl]benzamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | 著者 | Antanasijevic, A, Durst, M.A, Lavie, A, Caffrey, M. | | 登録日 | 2020-01-28 | | 公開日 | 2020-07-01 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of avian influenza hemagglutinin in complex with a small molecule entry inhibitor.
Life Sci Alliance, 3, 2020
|
|
6JCF
 
 | | Cryogenic structure of HIV-1 Integrase catalytic core domain by synchrotron | | 分子名称: | CACODYLATE ION, Integrase | | 著者 | Park, J.H, Han, J, Kim, T.H, Yun, J.H, Lee, W. | | 登録日 | 2019-01-28 | | 公開日 | 2019-07-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.153 Å) | | 主引用文献 | Non-Cryogenic Structure and Dynamics of HIV-1 Integrase Catalytic Core Domain by X-ray Free-Electron Lasers.
Int J Mol Sci, 20, 2019
|
|
6UIO
 
 | | Crystal structure of mouse CRES (Cystatin-Related Epididymal Spermatogenic) | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Cystatin-8, ... | | 著者 | Dominguez, M.J, Cornwall, G.A, Hewetson, A, Sutton, R.B. | | 登録日 | 2019-10-01 | | 公開日 | 2020-07-01 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Maturation of the functional mouse CRES amyloid from globular form.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6GYB
 
 | | Cryo-EM structure of the bacteria-killing type IV secretion system core complex from Xanthomonas citri | | 分子名称: | VirB10 protein, VirB7, VirB9 protein | | 著者 | Sgro, G.G, Costa, T.R.D, Farah, C.S, Waksman, G. | | 登録日 | 2018-06-28 | | 公開日 | 2018-10-24 | | 最終更新日 | 2019-12-11 | | 実験手法 | ELECTRON MICROSCOPY (3.28 Å) | | 主引用文献 | Cryo-EM structure of the bacteria-killing type IV secretion system core complex from Xanthomonas citri.
Nat Microbiol, 3, 2018
|
|
6GSC
 
 | | Sphingobacterium sp. T2 manganese superoxide dismutase catalyses the oxidative demethylation of polymeric lignin via generation of hydroxyl radical | | 分子名称: | MANGANESE (II) ION, Superoxide dismutase | | 著者 | Rashid, G.M.M, Zhang, X, Wilkinson, R.C, Fulop, V, Cottyn, B, Baumberger, S, Bugg, T.D.H. | | 登録日 | 2018-06-13 | | 公開日 | 2018-10-03 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | Sphingobacterium sp. T2 Manganese Superoxide Dismutase Catalyzes the Oxidative Demethylation of Polymeric Lignin via Generation of Hydroxyl Radical.
ACS Chem. Biol., 13, 2018
|
|
6HKS
 
 | |
5KDG
 
 | | Crystal Structure of Salmonella Typhimurium Effector GtgE | | 分子名称: | GLYCEROL, Gifsy-2 prophage protein, SULFATE ION | | 著者 | Kozlov, G, Xu, C, Wong, K, Gehring, K, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | 登録日 | 2016-06-08 | | 公開日 | 2016-11-16 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Crystal Structure of the Salmonella Typhimurium Effector GtgE.
PLoS ONE, 11, 2016
|
|
6VRX
 
 | |
6VJB
 
 | |
6NTY
 
 | | 2.1 A resolution structure of the Musashi-2 (Msi2) RNA recognition motif 1 (RRM1) domain | | 分子名称: | PHOSPHATE ION, RNA-binding protein Musashi homolog 2 | | 著者 | Lovell, S, Kashipathy, M.M, Battaile, K.P, Lan, L, Xiaoqing, W, Cooper, A, Gao, F.P, Xu, L. | | 登録日 | 2019-01-30 | | 公開日 | 2019-10-23 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal and solution structures of human oncoprotein Musashi-2 N-terminal RNA recognition motif 1.
Proteins, 88, 2020
|
|
5ZCK
 
 | | Structure of the RIP3 core region | | 分子名称: | SODIUM ION, peptide from Receptor-interacting serine/threonine-protein kinase 3 | | 著者 | Li, J, Wu, H. | | 登録日 | 2018-02-18 | | 公開日 | 2018-04-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.271 Å) | | 主引用文献 | The Structure of the Necrosome RIPK1-RIPK3 Core, a Human Hetero-Amyloid Signaling Complex.
Cell, 173, 2018
|
|
