2RN5
 
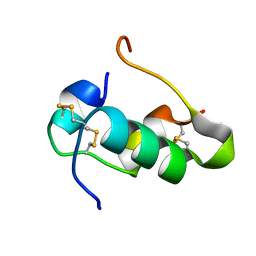 | | Humal Insulin Mutant B31Lys-B32Arg | | 分子名称: | Insulin | | 著者 | Bocian, W, Kozerski, L. | | 登録日 | 2007-12-06 | | 公開日 | 2008-10-28 | | 最終更新日 | 2022-03-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of biosynthetic engineered human insulin monomer B31(Lys)-B32(Arg) in water/acetonitrile solution. Comparison with the solution structure of native human insulin monomer
Biopolymers, 89, 2008
|
|
6K59
 
 | |
5CVW
 
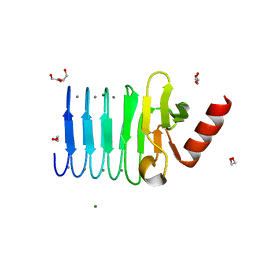 | | CRYSTAL STRUCTURE OF RTX DOMAIN BLOCK V OF ADENYLATE CYCLASE TOXIN FROM BORDETELLA PERTUSSIS | | 分子名称: | 1,2-ETHANEDIOL, Bifunctional hemolysin/adenylate cyclase, CALCIUM ION, ... | | 著者 | Motlova, L, Barinka, C, Bumba, L. | | 登録日 | 2015-07-27 | | 公開日 | 2015-09-09 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Calcium-Driven Folding of RTX Domain beta-Rolls Ratchets Translocation of RTX Proteins through Type I Secretion Ducts.
Mol.Cell, 62, 2016
|
|
5CXL
 
 | |
2R0H
 
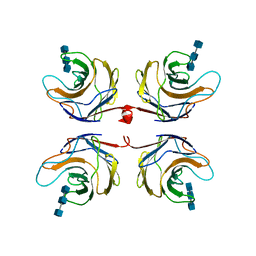 | | Fungal lectin CGL3 in complex with chitotriose (chitotetraose) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CGL3 lectin | | 著者 | Waelti, M.A, Walser, P.J, Thore, S, Gruenler, A, Ban, N, Kuenzler, M, Aebi, M. | | 登録日 | 2007-08-20 | | 公開日 | 2008-05-20 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural Basis for Chitotetraose Coordination by CGL3, a Novel Galectin-Related Protein from Coprinopsis cinerea
J.Mol.Biol., 379, 2008
|
|
2A3L
 
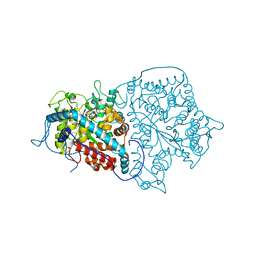 | | X-Ray Structure of Adenosine 5'-Monophosphate Deaminase from Arabidopsis Thaliana in Complex with Coformycin 5'-Phosphate | | 分子名称: | AMP deaminase, COFORMYCIN 5'-PHOSPHATE, PHOSPHATE ION, ... | | 著者 | Han, B.W, Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | 登録日 | 2005-06-25 | | 公開日 | 2005-07-19 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.34 Å) | | 主引用文献 | Membrane association, mechanism of action, and structure of Arabidopsis embryonic factor 1 (FAC1).
J.Biol.Chem., 281, 2006
|
|
5DNA
 
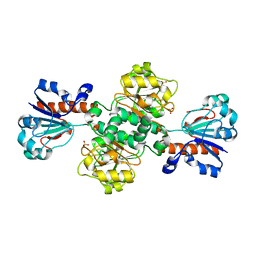 | | Crystal structure of Candida boidinii formate dehydrogenase | | 分子名称: | FORMATE DEHYDROGENASE, SULFATE ION | | 著者 | Guo, Q, Gakhar, L, Wichersham, K, Francis, K, Vardi-Kilshtain, A, Major, D.T, Cheatum, C.M, Kohen, A. | | 登録日 | 2015-09-09 | | 公開日 | 2016-05-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural and Kinetic Studies of Formate Dehydrogenase from Candida boidinii.
Biochemistry, 55, 2016
|
|
6DNA
 
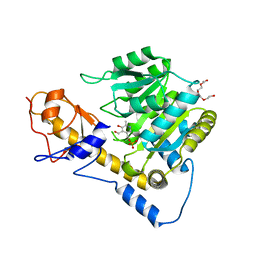 | | Crystal structure of T110A mutant human Glutamate oxaloacetate transaminase 1 (GOT1) | | 分子名称: | Aspartate aminotransferase, cytoplasmic, PYRIDOXAL-5'-PHOSPHATE, ... | | 著者 | Assar, Z, Holt, M.C, Stein, A.J, Lairson, L, Lyssiotis, C.A. | | 登録日 | 2018-06-06 | | 公開日 | 2018-11-14 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Biochemical Characterization and Structure-Based Mutational Analysis Provide Insight into the Binding and Mechanism of Action of Novel Aspartate Aminotransferase Inhibitors.
Biochemistry, 57, 2018
|
|
7DNA
 
 | |
1DNA
 
 | | D221(169)N MUTANT DOES NOT PROMOTE OPENING OF THE COFACTOR IMIDAZOLIDINE RING | | 分子名称: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | 著者 | Sage, C.R, Michelitsch, M.D, Finer-Moore, J, Stroud, R.M. | | 登録日 | 1998-06-25 | | 公開日 | 1998-11-04 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | D221 in thymidylate synthase controls conformation change, and thereby opening of the imidazolidine.
Biochemistry, 37, 1998
|
|
3DNA
 
 | |
4DNA
 
 | | CRYSTAL STRUCTURE OF putative glutathione reductase from Sinorhizobium meliloti 1021 | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Probable glutathione reductase | | 著者 | Malashkevich, V.N, Bhosle, R, Toro, R, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2012-02-08 | | 公開日 | 2012-03-21 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | CRYSTAL STRUCTURE OF putative glutathione reductase from Sinorhizobium meliloti 1021
To be Published
|
|
8DNA
 
 | |
2DKV
 
 | | Crystal structure of class I chitinase from Oryza sativa L. japonica | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, chitinase | | 著者 | Kezuka, Y, Nishizawa, Y, Watanabe, T, Nonaka, T. | | 登録日 | 2006-04-14 | | 公開日 | 2007-05-01 | | 最終更新日 | 2020-01-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of full-length class I chitinase from rice revealed by X-ray crystallography and small-angle X-ray scattering.
Proteins, 78, 2010
|
|
7MEI
 
 | | Composite structure of EC+EC | | 分子名称: | DNA (74-MER), DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | 著者 | Yang, C, Murakami, K. | | 登録日 | 2021-04-06 | | 公開日 | 2022-03-02 | | 実験手法 | ELECTRON MICROSCOPY (3.54 Å) | | 主引用文献 | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7MKA
 
 | | Structure of EC+EC (leading EC-focused) | | 分子名称: | DNA (40-MER), DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | 著者 | Yang, C, Murakami, K. | | 登録日 | 2021-04-22 | | 公開日 | 2022-04-27 | | 最終更新日 | 2023-05-17 | | 実験手法 | ELECTRON MICROSCOPY (3.54 Å) | | 主引用文献 | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7MK9
 
 | |
1WCM
 
 | | Complete 12-Subunit RNA Polymerase II at 3.8 Angstrom | | 分子名称: | DNA-DIRECTED RNA POLYMERASE II 13.6 KDA POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II 14.2 KDA POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II 19 KD POLYPEPTIDE, ... | | 著者 | Armache, K.-J, Mitterweger, S, Meinhart, A, Cramer, P. | | 登録日 | 2004-11-17 | | 公開日 | 2004-12-14 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Structures of Complete RNA Polymerase II and its Subcomplex,Rpb4/7
J.Biol.Chem., 280, 2005
|
|
7DN3
 
 | | Structure of Human RNA Polymerase III elongation complex | | 分子名称: | DNA (5'-D(P*TP*CP*GP*TP*CP*TP*GP*AP*TP*CP*TP*CP*GP*GP*AP*A)-3'), DNA (5'-D(P*TP*TP*CP*CP*GP*AP*GP*AP*TP*CP*AP*GP*AP*CP*GP*AP*GP*AP*TP*CP*GP*GP*G)-3'), DNA-directed RNA polymerase III subunit RPC1, ... | | 著者 | Li, L, Yu, Z, Zhao, D, Ren, Y, Hou, H, Xu, Y. | | 登録日 | 2020-12-08 | | 公開日 | 2021-03-17 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structure of human RNA polymerase III elongation complex.
Cell Res., 31, 2021
|
|
6XAV
 
 | | CryoEM Structure of E. coli Rho-dependent Transcription Pre-termination Complex bound with NusG | | 分子名称: | DNA (29-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Hao, Z.T, Kim, H.K, Walz, T, Nudler, E. | | 登録日 | 2020-06-04 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (7.7 Å) | | 主引用文献 | Pre-termination Transcription Complex: Structure and Function.
Mol.Cell, 81, 2021
|
|
8HKC
 
 | | Cryo-EM structure of E. coli RNAP sigma32 complex | | 分子名称: | DNA (54-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Wu, S, Ma, L.X. | | 登録日 | 2022-11-25 | | 公開日 | 2023-05-31 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.49 Å) | | 主引用文献 | Structural Insight into the Mechanism of sigma 32-Mediated Transcription Initiation of Bacterial RNA Polymerase.
Biomolecules, 13, 2023
|
|
6XAS
 
 | | CryoEM Structure of E. coli Rho-dependent Transcription Pre-termination Complex | | 分子名称: | DNA (29-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Hao, Z.T, Kim, H.K, Walz, T, Nudler, E. | | 登録日 | 2020-06-04 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Pre-termination Transcription Complex: Structure and Function.
Mol.Cell, 81, 2021
|
|
8GZG
 
 | |
6DRD
 
 | | RNA Pol II(G) | | 分子名称: | DNA-directed RNA polymerase II subunit GRINL1A, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11-a, ... | | 著者 | Yu, X, Jishage, M, Shi, Y, Ganesan, S, Sali, A, Chait, B.T, Asturias, F, Roeder, R.G. | | 登録日 | 2018-06-11 | | 公開日 | 2019-06-12 | | 最終更新日 | 2019-12-04 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Architecture of Pol II(G) and molecular mechanism of transcription regulation by Gdown1.
Nat. Struct. Mol. Biol., 25, 2018
|
|
8S5N
 
 | | RNA polymerase II core initially transcribing complex with an ordered RNA of 12 nt | | 分子名称: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | 著者 | Zhan, Y, Grabbe, F, Oberbeckmann, E, Dienemann, C, Cramer, P. | | 登録日 | 2024-02-24 | | 公開日 | 2024-04-10 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Three-step mechanism of promoter escape by RNA polymerase II.
Mol.Cell, 84, 2024
|
|
