7MU5
 
 | | Human DCTPP1 bound to Triptolide | | 分子名称: | MAGNESIUM ION, dCTP pyrophosphatase 1, triptolide | | 著者 | Hauk, G, Berger, J.M. | | 登録日 | 2021-05-14 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Triptolide sensitizes cancer cells to nucleoside DNA methyltransferase inhibitors through inhibition of DCTPP1 mediated cell-intrinsic resistance
To Be Published
|
|
7AKW
 
 | | Crystal structure of the viral rhodopsins chimera O1O2 | | 分子名称: | EICOSANE, RETINAL, chimera of viral rhodopsins OLPVR1 and OLPVRII | | 著者 | Kovalev, K, Zabelskii, D, Alekseev, A, Astashkin, R, Gordeliy, V. | | 登録日 | 2020-10-02 | | 公開日 | 2020-11-25 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Viral rhodopsins 1 are an unique family of light-gated cation channels.
Nat Commun, 11, 2020
|
|
7AKY
 
 | | Crystal structure of the viral rhodopsin OLPVR1 in P21212 space group | | 分子名称: | (2S)-2,3-dihydroxypropyl (9Z)-hexadec-9-enoate, EICOSANE, viral rhodopsin OLPVR1 | | 著者 | Kovalev, K, Zabelskii, D, Alekseev, A, Astashkin, R, Gordeliy, V. | | 登録日 | 2020-10-02 | | 公開日 | 2020-11-25 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Viral rhodopsins 1 are an unique family of light-gated cation channels.
Nat Commun, 11, 2020
|
|
7AKX
 
 | | Crystal structure of the viral rhodopsin OLPVR1 in P1 space group | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, OLEIC ACID, ... | | 著者 | Kovalev, K, Zabelskii, D, Alekseev, A, Astashkin, R, Gordeliy, V. | | 登録日 | 2020-10-02 | | 公開日 | 2020-11-25 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Viral rhodopsins 1 are an unique family of light-gated cation channels.
Nat Commun, 11, 2020
|
|
7EDB
 
 | |
5HGO
 
 | |
7E8I
 
 | | Structural insight into BRCA1-BARD1 complex recruitment to damaged chromatin | | 分子名称: | BRCA1-associated RING domain protein 1, DNA (145-MER), Histone H2A, ... | | 著者 | Dai, Y, Dai, L, Zhou, Z. | | 登録日 | 2021-03-01 | | 公開日 | 2021-06-30 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural insight into BRCA1-BARD1 complex recruitment to damaged chromatin.
Mol.Cell, 81, 2021
|
|
5HGK
 
 | |
5D7D
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 7-propyl-3-[2-(pyridin-3-yl)-1,3-thiazol-5-yl]-1,7-dihydro-6H-pyrazolo[3,4-b]pyridin-6-one, CHLORIDE ION, ... | | 著者 | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | 登録日 | 2015-08-13 | | 公開日 | 2015-11-11 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Discovery of Indazole Derivatives as a Novel Class of Bacterial Gyrase B Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
5HGM
 
 | |
1HT7
 
 | |
5HGN
 
 | |
5HGL
 
 | | Hexameric HIV-1 CA, open conformation | | 分子名称: | CHLORIDE ION, Capsid protein P24, N-METHYL-NALPHA-[(2-METHYL-1H-INDOL-3-YL)ACETYL]-N-PHENYL-L-PHENYLALANINAMIDE | | 著者 | Price, A.J, Jacques, D.A, James, L.C. | | 登録日 | 2016-01-08 | | 公開日 | 2016-08-10 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | HIV-1 uses dynamic capsid pores to import nucleotides and fuel encapsidated DNA synthesis.
Nature, 536, 2016
|
|
5D7R
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-hydroxy-5-[5-(6-hydroxy-7-propyl-2H-indazol-3-yl)-1,3-thiazol-2-yl]pyridine-2-carboxylic acid, CHLORIDE ION, ... | | 著者 | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | 登録日 | 2015-08-14 | | 公開日 | 2015-11-18 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Discovery of Indazole Derivatives as a Novel Class of Bacterial Gyrase B Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
1K8J
 
 | | NMR STRUCTURE OF THE CK14 DNA DUPLEX: A PORTION OF THE KNOWN NF-kB SEQUENCE CK1 | | 分子名称: | FIRST STRAND OF CK14 DNA DUPLEX, SECOND STRAND OF CK14 DNA DUPLEX | | 著者 | Volk, D.E, Yang, X, Fennewald, S.M, King, D.J, Bassett, S.E, Venkitachalam, S, Herzog, N, Luxon, B.A, Gorenstein, D.G. | | 登録日 | 2001-10-24 | | 公開日 | 2003-04-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure and design of dithiophosphate backbone aptamers targeting transcription factor NF-kappaB
Bioorg.Chem., 30, 2002
|
|
1OYW
 
 | |
1OYY
 
 | | Structure of the RecQ Catalytic Core bound to ATP-gamma-S | | 分子名称: | ATP-dependent DNA helicase, MANGANESE (II) ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Bernstein, D.A, Zittel, M.C, Keck, J.L. | | 登録日 | 2003-04-07 | | 公開日 | 2003-10-07 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | High-resolution structure of the E. coli RecQ helicase catalytic core
Embo J., 22, 2003
|
|
5HGP
 
 | |
4CYB
 
 | | DpsC from Streptomyces coelicolor | | 分子名称: | FE (III) ION, PUTATIVE DNA PROTECTION PROTEIN, SODIUM ION | | 著者 | Townsend, P.D, Hitchings, M.D, Del Sol, R, Pohl, E. | | 登録日 | 2014-04-10 | | 公開日 | 2014-06-25 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | A Tale of Tails: Deciphering the Contribution of Terminal Tails to the Biochemical Properties of Two Dps Proteins from Streptomyces Coelicolor
Cell.Mol.Life Sci., 71, 2014
|
|
4BAE
 
 | | Optimisation of pyrroleamides as mycobacterial GyrB ATPase inhibitors: Structure Activity Relationship and in vivo efficacy in the mouse model of tuberculosis | | 分子名称: | 2-[(3S,4R)-4-[(3-bromanyl-4-chloranyl-5-methyl-1H-pyrrol-2-yl)carbonylamino]-3-methoxy-piperidin-1-yl]-4-(2-methyl-1,2,4-triazol-3-yl)-1,3-thiazole-5-carboxylic acid, CALCIUM ION, DNA GYRASE SUBUNIT B, ... | | 著者 | Read, J.A, Gingell, H.G, Madhavapeddi, P. | | 登録日 | 2012-09-13 | | 公開日 | 2013-10-30 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Optimization of Pyrrolamides as Mycobacterial Gyrb ATPase Inhibitors: Structure Activity Relationship and in Vivo Efficacy in the Mouse Model of Tuberculosis.
Antimicrob.Agents Chemother., 58, 2014
|
|
4DA2
 
 | |
2ETX
 
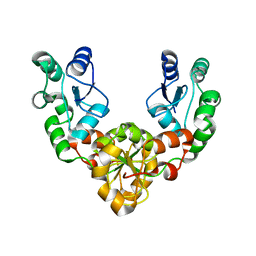 | | Crystal Structure of MDC1 Tandem BRCT Domains | | 分子名称: | Mediator of DNA damage checkpoint protein 1 | | 著者 | Wasielewski, E, Kim, Y, Joachimiak, A, Thompson, J.R, Mer, G. | | 登録日 | 2005-10-27 | | 公開日 | 2005-11-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.33 Å) | | 主引用文献 | Molecular Basis for the Association of Microcephalin (MCPH1) Protein with the Cell Division Cycle Protein 27 (Cdc27) Subunit of the Anaphase-promoting Complex.
J.Biol.Chem., 287, 2012
|
|
1KG6
 
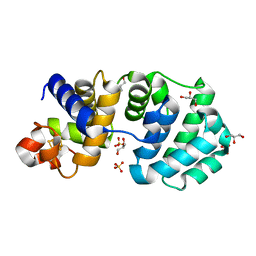 | | Crystal structure of the K142R mutant of E.coli MutY (core fragment) | | 分子名称: | A/G-specific adenine glycosylase, GLYCEROL, IRON/SULFUR CLUSTER, ... | | 著者 | Gilboa, R, Kilshtein, A, Zharkov, D.O, Kycia, J.H, Gerchman, S.E, Grollman, A.P, Shoham, G. | | 登録日 | 2001-11-26 | | 公開日 | 2002-11-26 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Analysis of the E.coli MutY DNA glycosylase structure and function by site-directed mutagenesis
To be Published
|
|
1KG4
 
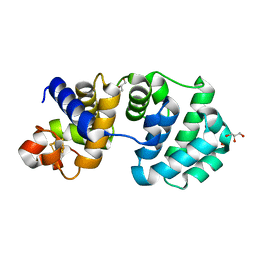 | | Crystal structure of the K142A mutant of E. coli MutY (core fragment) | | 分子名称: | A/G-specific adenine glycosylase, GLYCEROL, IRON/SULFUR CLUSTER, ... | | 著者 | Gilboa, R, Kilshtein, A, Zharkov, D.O, Kycia, J.H, Gerchman, S.E, Grollman, A.P, Shoham, G. | | 登録日 | 2001-11-26 | | 公開日 | 2002-11-26 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Analysis of the E.coli MutY DNA glycosylase structure and function by site-directed mutagenesis
To be Published
|
|
1KG3
 
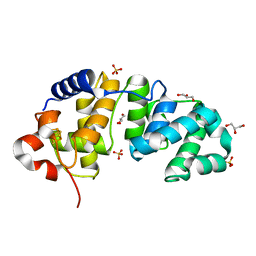 | | Crystal structure of the core fragment of MutY from E.coli at 1.55A resolution | | 分子名称: | A/G-specific adenine glycosylase, GLYCEROL, IRON/SULFUR CLUSTER, ... | | 著者 | Gilboa, R, Kilshtein, A, Zharkov, D.O, Kycia, J.H, Gerchman, S.E, Grollman, A.P, Shoham, G. | | 登録日 | 2001-11-26 | | 公開日 | 2002-11-26 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Analysis of the E.coli MutY DNA glycosylase structure and function by site-directed mutagenesis
To be Published
|
|
