4ZCW
 
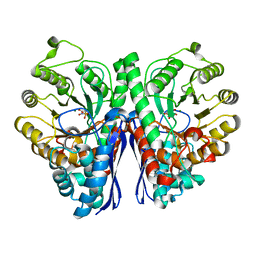 | | Structure of Human Enolase 2 in complex with SF2312 | | 分子名称: | Gamma-enolase, MAGNESIUM ION, [(3S,5S)-1,5-dihydroxy-2-oxopyrrolidin-3-yl]phosphonic acid | | 著者 | Leonard, P.G, Maxwell, D, Czako, B, Muller, F.L. | | 登録日 | 2015-04-16 | | 公開日 | 2016-07-20 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.992 Å) | | 主引用文献 | SF2312 is a natural phosphonate inhibitor of enolase.
Nat.Chem.Biol., 12, 2016
|
|
7XIT
 
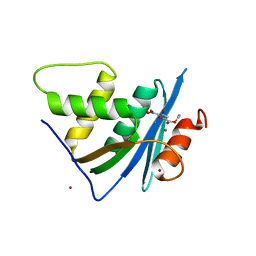 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with nitrofuran methoxy(methoxycarbonyl)phenyl ester | | 分子名称: | MANGANESE (II) ION, Reverse Transcriptase RNase H domain, ZINC ION, ... | | 著者 | Lu, H, Komukai, Y, Usami, K, Guo, Y, Qiao, X, Nukaga, M, Hoshino, T. | | 登録日 | 2022-04-14 | | 公開日 | 2022-04-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Computational and Crystallographic Analysis of Binding Structures of Inhibitory Compounds for HIV-1 RNase H Activity.
J.Chem.Inf.Model., 62, 2022
|
|
6B85
 
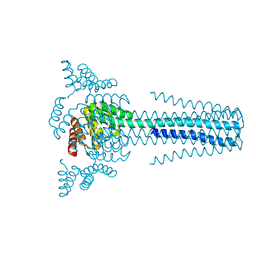 | | Crystal structure of transmembrane protein TMHC4_R | | 分子名称: | TMHC4_R | | 著者 | Lu, P, DiMaio, F, Min, D, Bowie, J, Wei, K.Y, Baker, D. | | 登録日 | 2017-10-05 | | 公開日 | 2018-03-14 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.889 Å) | | 主引用文献 | Accurate computational design of multipass transmembrane proteins.
Science, 359, 2018
|
|
6P8U
 
 | |
7XJ7
 
 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with nitrofuran methoxy(methoxycarbonyl)phenyl ester | | 分子名称: | MANGANESE (II) ION, Reverse Transcriptase RNase H domain, ZINC ION, ... | | 著者 | Lu, H, Komukai, Y, Usami, K, Guo, Y, Qiao, X, Nukaga, M, Hoshino, T. | | 登録日 | 2022-04-15 | | 公開日 | 2022-04-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Computational and Crystallographic Analysis of Binding Structures of Inhibitory Compounds for HIV-1 RNase H Activity.
J.Chem.Inf.Model., 62, 2022
|
|
7OA5
 
 | | RUVA COMPLEXED TO A HOLLIDAY JUNCTION. | | 分子名称: | CALCIUM ION, DNA (5'-D(*AP*GP*TP*TP*CP*GP*CP*GP*AP*GP*TP*TP*CP*GP*C)-3'), DNA (5'-D(*AP*GP*TP*TP*CP*GP*CP*GP*CP*GP*CP*GP*AP*AP*CP*T)-3'), ... | | 著者 | Roe, S.M, Pearl, L.H. | | 登録日 | 2021-04-19 | | 公開日 | 2021-04-28 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.378 Å) | | 主引用文献 | Crystal structure of an octameric RuvA-Holliday junction complex
Molecular Cell, 2, 1998
|
|
6BG8
 
 | |
8PJV
 
 | |
8PT8
 
 | | JNK1 covalently bound to RU135 cyclohexenone based inhibitor | | 分子名称: | GLYCEROL, Mitogen-activated protein kinase 8, methyl (1R,3R)-1-methyl-3-[[3-[[3-methyl-4-[(4-pyridin-3-ylpyrimidin-2-yl)amino]phenyl]carbamoyl]phenyl]methylcarbamoyl]-4-oxidanylidene-cyclohexane-1-carboxylate | | 著者 | Sok, P, Poti, A, Remenyi, A. | | 登録日 | 2023-07-13 | | 公開日 | 2024-07-24 | | 実験手法 | X-RAY DIFFRACTION (2.78 Å) | | 主引用文献 | Tunable c-Jun N-terminal kinase (JNK) inhibitors that target a specific cysteine by a reversible covalent bond
To Be Published
|
|
4ZDC
 
 | | Yeast enoyl-CoA isomerase complexed with octanoyl-CoA | | 分子名称: | 3,2-trans-enoyl-CoA isomerase, GLYCEROL, OCTANOYL-COENZYME A, ... | | 著者 | Onwukwe, G.U, Koski, M.K, Wierenga, R.K. | | 登録日 | 2015-04-17 | | 公開日 | 2015-11-11 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Structures of yeast peroxisomal Delta (3), Delta (2)-enoyl-CoA isomerase complexed with acyl-CoA substrate analogues: the importance of hydrogen-bond networks for the reactivity of the catalytic base and the oxyanion hole.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8PM5
 
 | |
7TIA
 
 | | Crystal structure of SARS-CoV-2 3CL in complex with inhibitor NK01-14 | | 分子名称: | 3C-like proteinase nsp5, THIOCYANATE ION, benzyl [(2S)-3-cyclopropyl-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-1-oxopropan-2-yl]carbamate | | 著者 | Forouhar, F, Liu, H, Iketani, S, Zack, A, Khanizeman, N, Bednarova, E, Fowler, B, Hong, S.J, Mohri, H, Nair, M.S, Huang, Y, Tay, N.E.S, Lee, S, Karan, C, Resnick, S.J, Quinn, C, Li, W, Shion, H, Jurtschenko, C, Lauber, M.A, McDonald, T, Stokes, M.E, Hurst, B, Rovis, T, Chavez, A, Ho, D.D, Stockwell, B.R. | | 登録日 | 2022-01-13 | | 公開日 | 2022-05-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19.
Nat Commun, 13, 2022
|
|
8PGC
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 2868 | | 分子名称: | 7-[(1~{S})-1-[5-[2-(1-azanylcyclopropyl)ethyl]-2-oxidanylidene-1,3-oxazol-3-yl]ethyl]-3-[3-fluoranyl-4-(methylsulfonylmethyl)phenyl]-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | 著者 | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | 登録日 | 2023-06-18 | | 公開日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Crystal structure of the metallo-beta-lactamase VIM1 with 2868
To Be Published
|
|
6MRJ
 
 | |
8PNE
 
 | |
7TIX
 
 | | Crystal structure of SARS-CoV-2 3CL in complex with inhibitor EB56 | | 分子名称: | 3C-like proteinase nsp5, MAGNESIUM ION, N~2~-{[(naphthalen-2-yl)methoxy]carbonyl}-N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | 著者 | Forouhar, F, Liu, H, Iketani, S, Zack, A, Khanizeman, N, Bednarova, E, Fowler, B, Hong, S.J, Mohri, H, Nair, M.S, Huang, Y, Tay, N.E.S, Lee, S, Karan, C, Resnick, S.J, Quinn, C, Li, W, Shion, H, Jurtschenko, C, Lauber, M.A, McDonald, T, Stokes, M.E, Hurst, B, Rovis, T, Chavez, A, Ho, D.D, Stockwell, B.R. | | 登録日 | 2022-01-14 | | 公開日 | 2022-05-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19.
Nat Commun, 13, 2022
|
|
7XJ4
 
 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with nitrofuran methoxy(methoxycarbonyl)phenyl ester | | 分子名称: | MANGANESE (II) ION, Reverse Transcriptase RNase H domain, S-[5-[(E)-2-phenylethenyl]-1,3,4-oxadiazol-2-yl] 5-nitrothiophene-2-carbothioate, ... | | 著者 | Lu, H, Komukai, Y, Usami, K, Guo, Y, Qiao, X, Nukaga, M, Hoshino, T. | | 登録日 | 2022-04-15 | | 公開日 | 2022-04-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Computational and Crystallographic Analysis of Binding Structures of Inhibitory Compounds for HIV-1 RNase H Activity.
J.Chem.Inf.Model., 62, 2022
|
|
6B1Y
 
 | | Crystal structure KPC-2 beta-lactamase complexed with WCK 5153 by co-crystallization | | 分子名称: | (2S,5Z)-1-formyl-5-imino-N'-[(3R)-1-(sulfooxy)pyrrolidine-3-carbonyl]piperidine-2-carbohydrazide, CHLORIDE ION, Carbapenem-hydrolyzing beta-lactamase KPC, ... | | 著者 | van den Akker, F, Nguyen, N.Q. | | 登録日 | 2017-09-19 | | 公開日 | 2018-08-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Strategic Approaches to Overcome Resistance against Gram-Negative Pathogens Using beta-Lactamase Inhibitors and beta-Lactam Enhancers: Activity of Three Novel Diazabicyclooctanes WCK 5153, Zidebactam (WCK 5107), and WCK 4234.
J. Med. Chem., 61, 2018
|
|
6P9F
 
 | | Crystal structure of RAR-related orphan receptor C (NHIS-RORGT(244-487)-L6-SRC1(678-692)) in complex with a phenyl (3-phenylpyrrolidin-3-yl)sulfone inhibitor | | 分子名称: | Nuclear receptor ROR-gamma, Nuclear receptor coactivator 1 chimera, trans-4-{(3R)-3-[(4-fluorophenyl)sulfonyl]-3-[4-(1,1,1,2,3,3,3-heptafluoropropan-2-yl)phenyl]pyrrolidine-1-carbonyl}cyclohexane-1-carboxylic acid | | 著者 | Sack, J. | | 登録日 | 2019-06-10 | | 公開日 | 2019-07-17 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Identification of potent, selective and orally bioavailable phenyl ((R)-3-phenylpyrrolidin-3-yl)sulfone analogues as ROR gamma t inverse agonists.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
8PGS
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 3380 | | 分子名称: | 7-[(1~{S})-1-[2-(aminomethyl)-6-oxidanylidene-5-oxa-7-azaspiro[3.4]octan-7-yl]ethyl]-3-(1-oxidanylidene-2,3-dihydroisoindol-5-yl)-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | 著者 | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | 登録日 | 2023-06-18 | | 公開日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.24 Å) | | 主引用文献 | Crystal structure of the metallo-beta-lactamase VIM1 with 3380
To Be Published
|
|
8E7O
 
 | |
6MRX
 
 | | Sialidase26 apo | | 分子名称: | Sialidase26 | | 著者 | Zaramela, L.S, Martino, C, Alisson-Silva, F, Rees, S.D, Diaz, S.L, Chuzel, L, Ganatra, M.B, Taron, C.H, Zuniga, C, Chang, G, Varki, A, Zengler, K. | | 登録日 | 2018-10-15 | | 公開日 | 2019-10-02 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Gut bacteria responding to dietary change encode sialidases that exhibit preference for red meat-associated carbohydrates.
Nat Microbiol, 4, 2019
|
|
4ZE2
 
 | | Saccharomyces cerevisiae CYP51 (Lanosterol 14-alpha demethylase) Y140H mutant complexed with itraconazole | | 分子名称: | 2-[(2R)-butan-2-yl]-4-{4-[4-(4-{[(2R,4S)-2-(2,4-dichlorophenyl)-2-(1H-1,2,4-triazol-1-ylmethyl)-1,3-dioxolan-4-yl]methoxy}phenyl)piperazin-1-yl]phenyl}-2,4-dihydro-3H-1,2,4-triazol-3-one, Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Sagatova, A, Keniya, M.V, Wilson, R, Monk, B.C, Tyndall, J.D.A. | | 登録日 | 2015-04-20 | | 公開日 | 2016-03-30 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Triazole resistance mediated by mutations of a conserved active site tyrosine in fungal lanosterol 14 alpha-demethylase.
Sci Rep, 6, 2016
|
|
8PH2
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 3746 | | 分子名称: | 7-[(1~{S})-1-[2-(aminomethyl)-6-oxidanylidene-5-oxa-7-azaspiro[3.4]octan-7-yl]ethyl]-3-[1-(2-azanylethyl)pyrazol-4-yl]-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | 著者 | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | 登録日 | 2023-06-18 | | 公開日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.13 Å) | | 主引用文献 | Crystal structure of the metallo-beta-lactamase VIM1 with 3746
To Be Published
|
|
6N6Q
 
 | |
