7BFK
 
 | | X-ray structure of SS-RNase-2 | | 分子名称: | Angiogenin-1 | | 著者 | Sica, F, Russo Krauss, I, Troisi, R. | | 登録日 | 2021-01-04 | | 公開日 | 2021-04-28 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | The structural features of an ancient ribonuclease from Salmo salar reveal an intriguing case of auto-inhibition.
Int.J.Biol.Macromol., 182, 2021
|
|
1LTA
 
 | | 2.2 ANGSTROMS CRYSTAL STRUCTURE OF E. COLI HEAT-LABILE ENTEROTOXIN (LT) WITH BOUND GALACTOSE | | 分子名称: | HEAT-LABILE ENTEROTOXIN, SUBUNIT A, SUBUNIT B, ... | | 著者 | Merritt, E.A, Sixma, T.K, Kalk, K.H, Van Zanten, B.A.M, Hol, W.G.J. | | 登録日 | 1993-09-15 | | 公開日 | 1994-01-31 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Galactose-binding site in Escherichia coli heat-labile enterotoxin (LT) and cholera toxin (CT).
Mol.Microbiol., 13, 1994
|
|
7WKC
 
 | |
7WXI
 
 | | GPR domain of Drosophila P5CS filament with glutamate and ATPgammaS | | 分子名称: | Delta-1-pyrroline-5-carboxylate synthase, GAMMA-GLUTAMYL PHOSPHATE | | 著者 | Liu, J.L, Zhong, J, Guo, C.J, Zhou, X. | | 登録日 | 2022-02-14 | | 公開日 | 2022-03-30 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Structural basis of dynamic P5CS filaments.
Elife, 11, 2022
|
|
1L9X
 
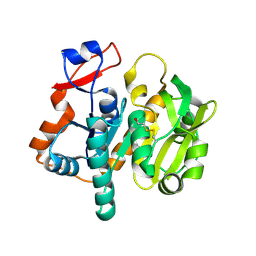 | | Structure of gamma-Glutamyl Hydrolase | | 分子名称: | BETA-MERCAPTOETHANOL, gamma-glutamyl hydrolase | | 著者 | Li, H, Ryan, T.J, Chave, K.J, Van Roey, P. | | 登録日 | 2002-03-26 | | 公開日 | 2002-04-10 | | 最終更新日 | 2021-04-07 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Three-dimensional structure of human gamma -glutamyl hydrolase. A class I glatamine amidotransferase adapted for a complex substate.
J.Biol.Chem., 277, 2002
|
|
7WXH
 
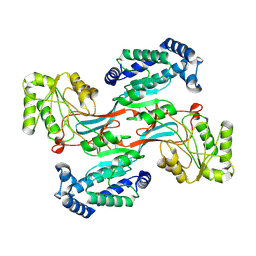 | | GPR domain open form of Drosophila P5CS filament with glutamate, ATP, and NADPH | | 分子名称: | Delta-1-pyrroline-5-carboxylate synthase | | 著者 | Liu, J.L, Zhong, J, Guo, C.J, Zhou, X. | | 登録日 | 2022-02-14 | | 公開日 | 2022-03-30 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structural basis of dynamic P5CS filaments.
Elife, 11, 2022
|
|
7WX4
 
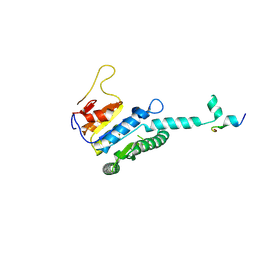 | |
7WXG
 
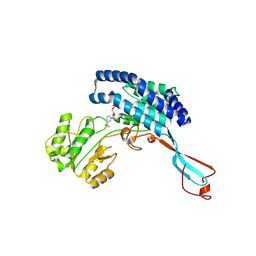 | | GPR domain closed form of Drosophila P5CS filament with glutamate, ATP, and NADPH | | 分子名称: | Delta-1-pyrroline-5-carboxylate synthase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Liu, J.L, Zhong, J, Guo, C.J, Zhou, X. | | 登録日 | 2022-02-14 | | 公開日 | 2022-03-30 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Structural basis of dynamic P5CS filaments.
Elife, 11, 2022
|
|
7WXF
 
 | |
7WX3
 
 | | GK domain of Drosophila P5CS filament with glutamate, ATP, and NADPH | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Delta-1-pyrroline-5-carboxylate synthase, GAMMA-GLUTAMYL PHOSPHATE, ... | | 著者 | Liu, J.L, Zhong, J, Guo, C.J, Zhou, X. | | 登録日 | 2022-02-14 | | 公開日 | 2022-04-06 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis of dynamic P5CS filaments.
Elife, 11, 2022
|
|
6DJQ
 
 | | Vps1 GTPase-BSE fusion complexed with GDP.AlF4- | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SODIUM ION, ... | | 著者 | Varlakhanova, N.V, Brady, T.M, Tornabene, B.A, Hosford, C.J, Chappie, J.S, Ford, M.G.J. | | 登録日 | 2018-05-25 | | 公開日 | 2018-08-22 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structures of the fungal dynamin-related protein Vps1 reveal a unique, open helical architecture.
J. Cell Biol., 217, 2018
|
|
2HNL
 
 | | Structure of the prostaglandin D synthase from the parasitic nematode Onchocerca volvulus | | 分子名称: | GLUTATHIONE, Glutathione S-transferase 1 | | 著者 | Perbandt, M, Hoppner, J, Betzel, C, Liebau, E. | | 登録日 | 2006-07-13 | | 公開日 | 2007-07-17 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of the extracellular glutathione S-transferase OvGST1 from the human pathogenic parasite Onchocerca volvulus.
J.Mol.Biol., 377, 2008
|
|
1KFC
 
 | | CRYSTAL STRUCTURE OF ALPHAT183V MUTANT OF TRYPTOPHAN SYNTHASE FROM SALMONELLA TYPHIMURIUM With Indole Propanol Phosphate | | 分子名称: | INDOLE-3-PROPANOL PHOSPHATE, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | 著者 | Kulik, V, Weyand, M, Siedel, R, Niks, D, Arac, D, Dunn, M.F, Schlichting, I. | | 登録日 | 2001-11-20 | | 公開日 | 2003-01-07 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | On the Role of AlphaTHR183 in the Allosteric Regulation and Catalytic Mechanism of Tryptophan Synthase
J.Mol.Biol., 324, 2002
|
|
6UEH
 
 | | Crystal structure of a ruminal GH26 endo-beta-1,4-mannanase | | 分子名称: | ACETATE ION, CALCIUM ION, Cow rumen GH26 endo-mannanase | | 著者 | Mandelli, F, Morais, M.A.B, Lima, E.A, Persinoti, G.F, Murakami, M.T. | | 登録日 | 2019-09-21 | | 公開日 | 2020-03-11 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.849 Å) | | 主引用文献 | Spatially remote motifs cooperatively affect substrate preference of a ruminal GH26-type endo-beta-1,4-mannanase.
J.Biol.Chem., 295, 2020
|
|
3RT3
 
 | |
1KFE
 
 | | CRYSTAL STRUCTURE OF ALPHAT183V MUTANT OF TRYPTOPHAN SYNTHASE FROM SALMONELLA TYPHIMURIUM WITH L-Ser Bound To The Beta Site | | 分子名称: | SODIUM ION, TRYPTOPHAN SYNTHASE ALPHA CHAIN, TRYPTOPHAN SYNTHASE BETA CHAIN, ... | | 著者 | Kulik, V, Weyand, M, Siedel, R, Niks, D, Arac, D, Dunn, M.F, Schlichting, I. | | 登録日 | 2001-11-20 | | 公開日 | 2003-01-07 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | On the Role of AlphaTHR183 in the Allosteric Regulation and Catalytic Mechanism of Tryptophan Synthase
J.Mol.Biol., 324, 2002
|
|
6DAE
 
 | |
1KQC
 
 | | Structure of Nitroreductase from E. cloacae Complex with Inhibitor Acetate | | 分子名称: | ACETATE ION, FLAVIN MONONUCLEOTIDE, OXYGEN-INSENSITIVE NAD(P)H NITROREDUCTASE | | 著者 | Haynes, C.A, Koder, R.L, Miller, A.F, Rodgers, D.W. | | 登録日 | 2002-01-04 | | 公開日 | 2002-02-13 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structures of nitroreductase in three states: effects of inhibitor binding and reduction.
J.Biol.Chem., 277, 2002
|
|
7CI0
 
 | | Microbial Hormone-sensitive lipase E53 mutant S162A | | 分子名称: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | 著者 | Yang, X, Li, Z, Xu, X, Li, J. | | 登録日 | 2020-07-06 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Mechanism and Structural Insights Into a Novel Esterase, E53, Isolated From Erythrobacter longus .
Front Microbiol, 12, 2021
|
|
7CIH
 
 | | Microbial Hormone-sensitive lipase E53 mutant S285G | | 分子名称: | (4-nitrophenyl) hexanoate, 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, ... | | 著者 | Yang, X, Li, Z, Xu, X, Li, J. | | 登録日 | 2020-07-07 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.789 Å) | | 主引用文献 | Mechanism and Structural Insights Into a Novel Esterase, E53, Isolated From Erythrobacter longus .
Front Microbiol, 12, 2021
|
|
6UGM
 
 | | Structural basis of COMPASS eCM recognition of an unmodified nucleosome | | 分子名称: | Bre2, DNA (146-MER), H3 N-terminus, ... | | 著者 | Hsu, P.L, Shi, H, Zheng, N. | | 登録日 | 2019-09-26 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural Basis of H2B Ubiquitination-Dependent H3K4 Methylation by COMPASS.
Mol.Cell, 76, 2019
|
|
2KYI
 
 | | Solution NMR structure of Dsy0195(21-82) protein from Desulfitobacterium Hafniense. Northeast Structural Genomics Consortium Target DhR8C | | 分子名称: | Uncharacterized protein | | 著者 | Yang, Y, Ramelot, T.A, Cort, J.R, Wang, H, Ciccosanti, C, Foote, E.L, Jiang, M, Janjua, H, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2010-05-27 | | 公開日 | 2010-06-09 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Combining NMR and EPR methods for homodimer protein structure determination.
J.Am.Chem.Soc., 132, 2010
|
|
2KSA
 
 | |
2KHS
 
 | | Solution structure of SNase121:SNase(111-143) complex | | 分子名称: | Nuclease, Thermonuclease | | 著者 | Geng, Y, Feng, Y, Xie, T, Shan, L, Wang, J. | | 登録日 | 2009-04-10 | | 公開日 | 2009-10-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The native-like interactions between SNase121 and SNase(111-143) fragments induce the recovery of their native-like structures and the ability to degrade DNA.
Biochemistry, 48, 2009
|
|
1II5
 
 | |
