7O81
 
 | | Rabbit 80S ribosome colliding in another ribosome stalled by the SARS-CoV-2 pseudoknot | | 分子名称: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | 著者 | Bhatt, P.R, Scaiola, A, Leibundgut, M.A, Atkins, J.F, Ban, N. | | 登録日 | 2021-04-14 | | 公開日 | 2021-06-02 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome.
Science, 372, 2021
|
|
7O7Z
 
 | | Rabbit 80S ribosome stalled close to the mutated SARS-CoV-2 slippery site by a pseudoknot (classified for pseudoknot) | | 分子名称: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | 著者 | Bhatt, P.R, Scaiola, A, Leibundgut, M.A, Atkins, J.F, Ban, N. | | 登録日 | 2021-04-14 | | 公開日 | 2021-06-02 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.4 Å) | | 主引用文献 | Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome.
Science, 372, 2021
|
|
7O7Y
 
 | | Rabbit 80S ribosome stalled close to the mutated SARS-CoV-2 slippery site by a pseudoknot (high resolution) | | 分子名称: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | 著者 | Bhatt, P.R, Scaiola, A, Leibundgut, M.A, Atkins, J.F, Ban, N. | | 登録日 | 2021-04-14 | | 公開日 | 2021-06-02 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.2 Å) | | 主引用文献 | Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome.
Science, 372, 2021
|
|
8GLP
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (Consensus LSU focused refined structure) | | 分子名称: | 1,4-DIAMINOBUTANE, 18S rRNA, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | 著者 | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Watson, Z.L, Altman, R.B, Blanchard, S.C. | | 登録日 | 2023-03-22 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (1.67 Å) | | 主引用文献 | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
4V97
 
 | | Crystal structure of the bacterial ribosome ram mutation G299A. | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Fagan, C.E, Dunkle, J.A, Maehigashi, T, Dunham, C.M. | | 登録日 | 2012-04-06 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (3.516 Å) | | 主引用文献 | Reorganization of an intersubunit bridge induced by disparate 16S ribosomal ambiguity mutations mimics an EF-Tu-bound state.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7OLC
 
 | | Thermophilic eukaryotic 80S ribosome at idle POST state | | 分子名称: | 18S rRNA, 26S rRNA, 40S ribosomal protein S0, ... | | 著者 | Kisonaite, M, Wild, K, Sinning, I. | | 登録日 | 2021-05-19 | | 公開日 | 2022-01-26 | | 最終更新日 | 2022-04-06 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | High-resolution structures of a thermophilic eukaryotic 80S ribosome reveal atomistic details of translocation.
Nat Commun, 13, 2022
|
|
6Z6L
 
 | | Cryo-EM structure of human CCDC124 bound to 80S ribosomes | | 分子名称: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | 著者 | Wells, J.N, Buschauer, R, Mackens-Kiani, T, Best, K, Kratzat, H, Berninghausen, O, Becker, T, Cheng, J, Beckmann, R. | | 登録日 | 2020-05-28 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structure and function of yeast Lso2 and human CCDC124 bound to hibernating ribosomes.
Plos Biol., 18, 2020
|
|
7OLD
 
 | | Thermophilic eukaryotic 80S ribosome at pe/E (TI)-POST state | | 分子名称: | 18S rRNA, 26S rRNA, 40S ribosomal protein S0, ... | | 著者 | Kisonaite, M, Wild, K, Sinning, I. | | 登録日 | 2021-05-19 | | 公開日 | 2022-01-26 | | 最終更新日 | 2022-02-09 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | High-resolution structures of a thermophilic eukaryotic 80S ribosome reveal atomistic details of translocation.
Nat Commun, 13, 2022
|
|
6CFJ
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with histidyl-CAM and bound to mRNA and A-, P-, and E-site tRNAs at 2.8A resolution | | 分子名称: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Tereshchenkov, A.G, Dobosz-Bartoszek, M, Osterman, I.A, Marks, J, Sergeeva, V.A, Kasatsky, P, Komarova, E.S, Stavrianidi, A.N, Rodin, I.A, Konevega, A.L, Sergiev, P.V, Sumbatyan, N.V, Mankin, A.S, Bogdanov, A.A, Polikanov, Y.S. | | 登録日 | 2018-02-15 | | 公開日 | 2018-03-07 | | 最終更新日 | 2025-03-19 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Binding and Action of Amino Acid Analogs of Chloramphenicol upon the Bacterial Ribosome.
J. Mol. Biol., 430, 2018
|
|
4V8J
 
 | | Crystal structure of the bacterial ribosome ram mutation G347U. | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Fagan, C.E, Dunkle, J.A, Maehigashi, T, Dunham, C.M. | | 登録日 | 2011-12-20 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (3.9 Å) | | 主引用文献 | Reorganization of an intersubunit bridge induced by disparate 16S ribosomal ambiguity mutations mimics an EF-Tu-bound state.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8P9A
 
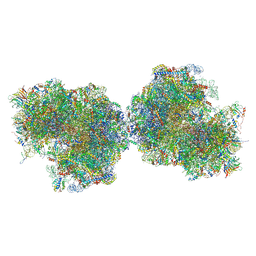 | | 80S yeast ribosome in complex with Methyllissoclimide | | 分子名称: | (3~{R})-3-[(1~{S})-2-[(1~{R},3~{S},4~{a}~{S},7~{S},8~{a}~{S})-5,5,7,8~{a}-tetramethyl-2-methylidene-3-oxidanyl-3,4,4~{a},6,7,8-hexahydro-1~{H}-naphthalen-1-yl]-1-oxidanyl-ethyl]pyrrolidine-2,5-dione, 16S ribosomal RNA, 25S ribosomal RNA, ... | | 著者 | Terrosu, S, Yusupov, M, Vanderwal, C. | | 登録日 | 2023-06-05 | | 公開日 | 2024-11-27 | | 最終更新日 | 2025-04-30 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Synthesis of Differentially Halogenated Lissoclimide Analogues To Probe Ribosome E-Site Binding.
Acs Chem.Biol., 20, 2025
|
|
3J7P
 
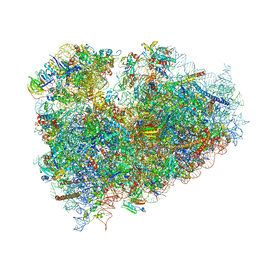 | | Structure of the 80S mammalian ribosome bound to eEF2 | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | 著者 | Voorhees, R.M, Fernandez, I.S, Scheres, S.H.W, Hegde, R.S. | | 登録日 | 2014-08-01 | | 公開日 | 2014-09-03 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structure of the Mammalian ribosome-sec61 complex to 3.4 a resolution.
Cell(Cambridge,Mass.), 157, 2014
|
|
6ICV
 
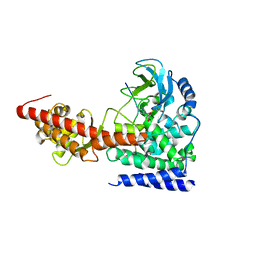 | | Structure of SETD3 bound to SAH and unmodified actin | | 分子名称: | Actin, cytoplasmic 1, Histone-lysine N-methyltransferase setd3, ... | | 著者 | Guo, Q, Liao, S, Xu, C, Structural Genomics Consortium (SGC) | | 登録日 | 2018-09-07 | | 公開日 | 2019-02-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structural insights into SETD3-mediated histidine methylation on beta-actin.
Elife, 8, 2019
|
|
6ICT
 
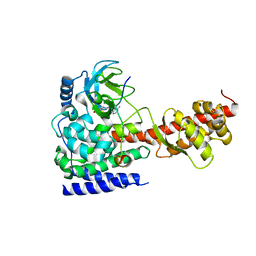 | | Structure of SETD3 bound to SAH and methylated actin | | 分子名称: | Actin, cytoplasmic 1, Histone-lysine N-methyltransferase setd3, ... | | 著者 | Guo, Q, Liao, S, Xu, C, Structural Genomics Consortium (SGC) | | 登録日 | 2018-09-07 | | 公開日 | 2019-02-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.952 Å) | | 主引用文献 | Structural insights into SETD3-mediated histidine methylation on beta-actin.
Elife, 8, 2019
|
|
6Y57
 
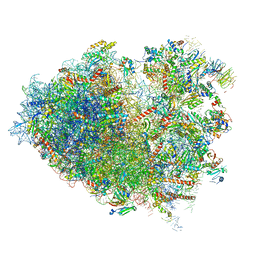 | | Structure of human ribosome in hybrid-PRE state | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | 著者 | Bhaskar, V, Schenk, A.D, Cavadini, S, von Loeffelholz, O, Natchiar, S.K, Klaholz, B.P, Chao, J.A. | | 登録日 | 2020-02-25 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Dynamics of uS19 C-Terminal Tail during the Translation Elongation Cycle in Human Ribosomes.
Cell Rep, 31, 2020
|
|
4UG0
 
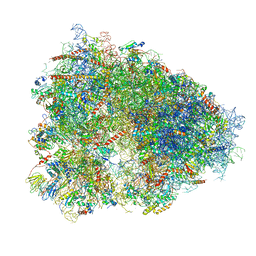 | | STRUCTURE OF THE HUMAN 80S RIBOSOME | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S RIBOSOMAL PROTEIN, ... | | 著者 | Khatter, H, Myasnikov, A.G, Natchiar, S.K, Klaholz, B.P. | | 登録日 | 2015-03-20 | | 公開日 | 2015-06-10 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structure of the human 80S ribosome
NATURE, 520, 2015
|
|
8JHF
 
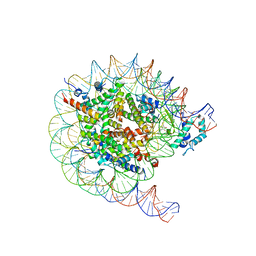 | | Native SUV420H1 bound to 167-bp nucleosome | | 分子名称: | DNA (160-MER), Histone H2A.Z, Histone H2B type 1-K, ... | | 著者 | Lin, F, Li, W. | | 登録日 | 2023-05-23 | | 公開日 | 2023-11-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (3.68 Å) | | 主引用文献 | Structural basis of nucleosomal H4K20 recognition and methylation by SUV420H1 methyltransferase.
Cell Discov, 9, 2023
|
|
8JHG
 
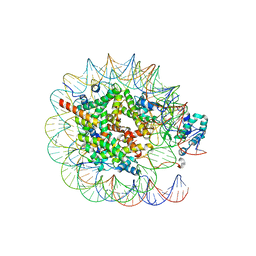 | | Native SUV420H1 bound to 167-bp nucleosome | | 分子名称: | DNA (160-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | 著者 | Lin, F, Li, W. | | 登録日 | 2023-05-23 | | 公開日 | 2023-11-22 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (3.58 Å) | | 主引用文献 | Structural basis of nucleosomal H4K20 recognition and methylation by SUV420H1 methyltransferase.
Cell Discov, 9, 2023
|
|
6Z6M
 
 | | Cryo-EM structure of human 80S ribosomes bound to EBP1, eEF2 and SERBP1 | | 分子名称: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | 著者 | Wells, J.N, Buschauer, R, Mackens-Kiani, T, Best, K, Kratzat, H, Berninghausen, O, Becker, T, Cheng, J, Beckmann, R. | | 登録日 | 2020-05-28 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structure and function of yeast Lso2 and human CCDC124 bound to hibernating ribosomes.
Plos Biol., 18, 2020
|
|
6HHQ
 
 | | Crystal structure of compound C45 bound to the yeast 80S ribosome | | 分子名称: | (3~{R})-3-[(1~{S})-2-[(1~{S},4~{a}~{R},6~{S},7~{S},8~{a}~{R})-6,7-bis(chloranyl)-5,5,8~{a}-trimethyl-2-methylidene-3,4,4~{a},6,7,8-hexahydro-1~{H}-naphthalen-1-yl]-1-oxidanyl-ethyl]pyrrolidine-2,5-dione, 18S ribosomal RNA, 25S ribosomal RNA, ... | | 著者 | Pellegrino, S, Vanderwal, C.D, Yusupov, M. | | 登録日 | 2018-08-28 | | 公開日 | 2019-02-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (3.10000038 Å) | | 主引用文献 | Understanding the role of intermolecular interactions between lissoclimides and the eukaryotic ribosome.
Nucleic Acids Res., 47, 2019
|
|
9O3L
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with macrolide erythromycin, mRNA, deacylated A-site tRNAphe, P-site fMRC-peptidyl-tRNAmet, and deacylated E-site tRNAphe at 2.75A resolution | | 分子名称: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Syroegin, E.A, Aleksandrova, E.V, Kruglov, A.A, Paranjpe, M.N, Svetlov, M.S, Polikanov, Y.S. | | 登録日 | 2025-04-07 | | 公開日 | 2025-06-25 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structural insights into context-specific inhibition of bacterial translation by macrolides.
Biorxiv, 2025
|
|
9O3K
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with macrolide erythromycin, mRNA, aminoacylated A-site Lys-tRNAlys, P-site fMAC-peptidyl-tRNAmet, and deacylated E-site tRNAlys at 2.70A resolution | | 分子名称: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Syroegin, E.A, Aleksandrova, E.V, Kruglov, A.A, Paranjpe, M.N, Svetlov, M.S, Polikanov, Y.S. | | 登録日 | 2025-04-07 | | 公開日 | 2025-06-25 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural insights into context-specific inhibition of bacterial translation by macrolides.
Biorxiv, 2025
|
|
9O3J
 
 | | Crystal structure of the wild-type drug-free Thermus thermophilus 70S ribosome in complex with mRNA, aminoacylated A-site Lys-tRNAlys, P-site fMRC-peptidyl-tRNAmet, and deacylated E-site tRNAlys at 2.60A resolution | | 分子名称: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Syroegin, E.A, Aleksandrova, E.V, Kruglov, A.A, Paranjpe, M.N, Svetlov, M.S, Polikanov, Y.S. | | 登録日 | 2025-04-07 | | 公開日 | 2025-06-25 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural insights into context-specific inhibition of bacterial translation by macrolides.
Biorxiv, 2025
|
|
9O3H
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with macrolide erythromycin, mRNA, aminoacylated A-site Lys-tRNAlys, P-site fMRC-peptidyl-tRNAmet, and deacylated E-site tRNAlys at 2.65A resolution | | 分子名称: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Syroegin, E.A, Aleksandrova, E.V, Kruglov, A.A, Paranjpe, M.N, Svetlov, M.S, Polikanov, Y.S. | | 登録日 | 2025-04-07 | | 公開日 | 2025-06-25 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structural insights into context-specific inhibition of bacterial translation by macrolides.
Biorxiv, 2025
|
|
4QEO
 
 | | crystal structure of KRYPTONITE in complex with mCHH DNA, H3(1-15) peptide and SAH | | 分子名称: | DNA 5'-ACTGATGAGTACCAT-3', DNA 5'-GGTACT(5CM)ATCAGTAT-3', Histone H3, ... | | 著者 | Du, J, Li, S, Patel, D.J. | | 登録日 | 2014-05-17 | | 公開日 | 2014-07-30 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Mechanism of DNA Methylation-Directed Histone Methylation by KRYPTONITE.
Mol.Cell, 55, 2014
|
|
