3DS9
 
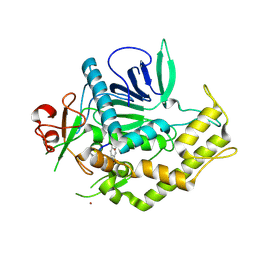 | |
3DSS
 
 | | Crystal structure of RabGGTase(DELTA LRR; DELTA IG) | | 分子名称: | CALCIUM ION, Geranylgeranyl transferase type-2 subunit alpha, Geranylgeranyl transferase type-2 subunit beta, ... | | 著者 | Guo, Z, Yu, S, Goody, R.S, Alexandrov, K, Blankenfeldt, W. | | 登録日 | 2008-07-14 | | 公開日 | 2008-09-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structures of RabGGTase-substrate/product complexes provide insights into the evolution of protein prenylation
Embo J., 27, 2008
|
|
3M9C
 
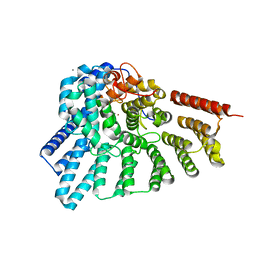
 | | Crystal structure of the membrane domain of respiratory complex I from Escherichia coli | | 分子名称: | NADH-quinone oxidoreductase subunit NuoL, NADH-quinone oxidoreductase subunit NuoM, NADH-quinone oxidoreductase subunit NuoN, ... | | 著者 | Efremov, R.G, Baradaran, R, Sazanov, L.A. | | 登録日 | 2010-03-22 | | 公開日 | 2010-05-26 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.9 Å) | | 主引用文献 | The architecture of respiratory complex I
Nature, 465, 2010
|
|
4KC5
 
 | |
3MDW
 
 | | The structure of N-formimino-L-Glutamate Iminohydrolase from Pseudomonas aeruginosa complexed with N-formimino-L-Aspartate | | 分子名称: | GLYCEROL, N-[(E)-iminomethyl]-L-aspartic acid, N-formimino-L-Glutamate Iminohydrolase, ... | | 著者 | Fedorov, A.A, Fedorov, E.V, Marti-Arbona, R, Raushel, F.M, Almo, S.C. | | 登録日 | 2010-03-30 | | 公開日 | 2011-03-09 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.8979 Å) | | 主引用文献 | Structure of N-Formimino-l-glutamate Iminohydrolase from Pseudomonas aeruginosa.
Biochemistry, 54, 2015
|
|
3DTG
 
 | |
4KEC
 
 | | SbHCT-complex form | | 分子名称: | (3R,4S,5R)-3,4-dihydroxy-5-{[(2E)-3-(4-hydroxyphenyl)prop-2-enoyl]oxy}cyclohex-1-ene-1-carboxylic acid, COENZYME A, Hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyl transferase | | 著者 | Walker, A.M, Hayes, R.P, Youn, B, Vermerris, W, Sattler, S.E, Kang, C. | | 登録日 | 2013-04-25 | | 公開日 | 2013-05-22 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Elucidation of the structure and reaction mechanism of sorghum hydroxycinnamoyltransferase and its structural relationship to other coenzyme a-dependent transferases and synthases.
Plant Physiol., 162, 2013
|
|
4KEK
 
 | |
3D8Z
 
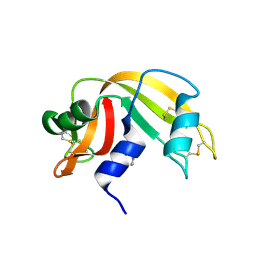 | | RNase A- 5'-Deoxy-5'-N-pyrrolidinothymidine complex | | 分子名称: | 1-(2,5-dideoxy-5-pyrrolidin-1-yl-beta-L-erythro-pentofuranosyl)-5-methylpyrimidine-2,4(1H,3H)-dione, CITRATE ANION, Ribonuclease pancreatic | | 著者 | Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G. | | 登録日 | 2008-05-26 | | 公開日 | 2009-02-10 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Morpholino, piperidino, and pyrrolidino derivatives of pyrimidine nucleosides as inhibitors of ribonuclease A: synthesis, biochemical, and crystallographic evaluation
J.Med.Chem., 52, 2009
|
|
4UCH
 
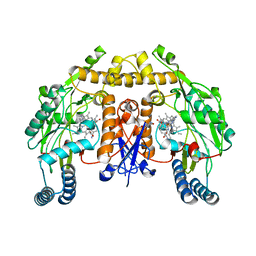 | | Structure of human nNOS R354A G357D mutant heme domain in complex with 3-(((2-((2-(1H-IMIDAZOL-1-YL)PYRIMIDIN-4-YL)ETHYL)AMINO)METHYL) BENZONITRILE | | 分子名称: | 3-[({2-[2-(1H-imidazol-1-yl)pyrimidin-4-yl]ethyl}amino)methyl]benzonitrile, 5,6,7,8-TETRAHYDROBIOPTERIN, NITRIC OXIDE SYNTHASE, ... | | 著者 | Li, H, Poulos, T.L. | | 登録日 | 2014-12-03 | | 公開日 | 2014-12-24 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Novel 2,4-Disubstituted Pyrimidines as Potent, Selective, and Cell-Permeable Inhibitors of Neuronal Nitric Oxide Synthase.
J.Med.Chem., 58, 2015
|
|
3D9S
 
 | | Human Aquaporin 5 (AQP5) - High Resolution X-ray Structure | | 分子名称: | Aquaporin-5, O-[(S)-{[(2S)-2-(hexanoyloxy)-3-(tetradecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-D-serine | | 著者 | Horsefield, R, Norden, K, Fellert, M, Backmark, A, Tornroth-Horsefield, S, Terwisscha Van Scheltinga, A.C, Kvassman, J, Kjellbom, P, Johanson, U, Neutze, R. | | 登録日 | 2008-05-27 | | 公開日 | 2008-08-26 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | High-resolution x-ray structure of human aquaporin 5
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3MAZ
 
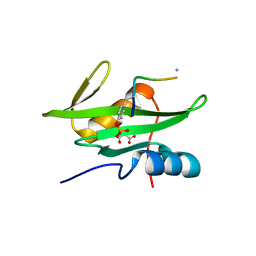 | | Crystal Structure of the Human BRDG1/STAP-1 SH2 Domain in Complex with the NTAL pTyr136 Peptide | | 分子名称: | CheD family protein, MALONATE ION, Signal-transducing adaptor protein 1 | | 著者 | Kaneko, T, Huang, H, Zhao, B, Li, L, Liu, H, Voss, C.K, Wu, C, Schiller, M.R, Li, S.S. | | 登録日 | 2010-03-24 | | 公開日 | 2010-05-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Loops govern SH2 domain specificity by controlling access to binding pockets.
Sci.Signal., 3, 2010
|
|
4UDP
 
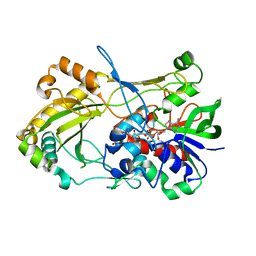 | | Crystal structure of 5-hydroxymethylfurfural oxidase (HMFO) in the oxidized state | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, GLUCOSE-METHANOL-CHOLINE OXIDOREDUCTASE | | 著者 | Dijkman, W, Binda, C, Fraaije, M, Mattevi, A. | | 登録日 | 2014-12-11 | | 公開日 | 2015-03-18 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure-Based Enzyme Tailoring of 5-Hydroxymethylfurfural Oxidase
Acs Catalysis, 5, 2015
|
|
3MB5
 
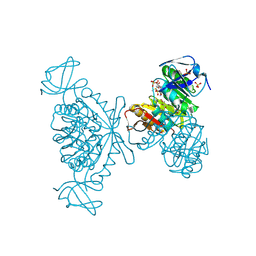 | |
4KFG
 
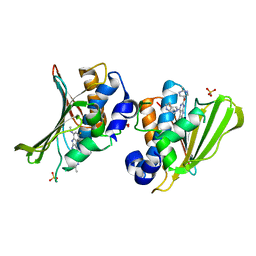 | | The DNA Gyrase B ATP binding domain of Escherichia coli in complex with a small molecule inhibitor. | | 分子名称: | 6-fluoro-4-[(3aR,6aR)-hexahydropyrrolo[3,4-b]pyrrol-5(1H)-yl]-N-methyl-2-[(2-methylpyrimidin-5-yl)oxy]-9H-pyrimido[4,5-b]indol-8-amine, DNA gyrase subunit B, SULFATE ION | | 著者 | Bensen, D.C, Akers-Rodriguez, S, Lam, T, Tari, L.W. | | 登録日 | 2013-04-26 | | 公開日 | 2014-01-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | A new class of type IIA topoisomerase inhibitors with broad-spectrum antibacterial activity
To be Published
|
|
3DR6
 
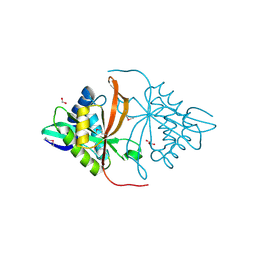 | | Structure of yncA, a putative ACETYLTRANSFERASE from Salmonella typhimurium | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, yncA | | 著者 | Singer, A.U, Skarina, T, Onopriyenko, O, Edwards, A.M, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2008-07-10 | | 公開日 | 2008-09-09 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Funded by the national institute of
allergy and infectious diseases of nih (contract number
hhsn272200700058c).
To be Published
|
|
4UDA
 
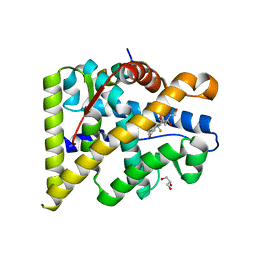 | | MR in complex with dexamethasone | | 分子名称: | DEXAMETHASONE, GLYCEROL, MINERALOCORTICOID RECEPTOR, ... | | 著者 | Edman, K, Hogner, A, Hussein, A, Bjursell, M, Aagaard, A, Backstrom, S, Bodin, C, Wissler, L, Jellesmark-Jensen, T, Cavallin, A, Karlsson, U, Nilsson, E, Lecina, D, Takahashi, R, Grebner, C, Lepisto, M, Guallar, V. | | 登録日 | 2014-12-09 | | 公開日 | 2015-11-25 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Ligand Binding Mechanism in Steroid Receptors: From Conserved Plasticity to Differential Evolutionary Constraints.
Structure, 23, 2015
|
|
4KFU
 
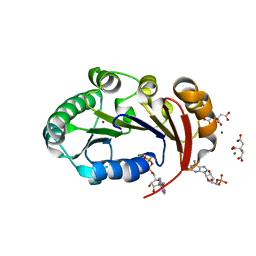 | | Structure of the genome packaging NTPase B204 from Sulfolobus turreted icosahedral virus 2 in complex with AMPPCP | | 分子名称: | CITRATE ANION, Genome packaging NTPase B204, MAGNESIUM ION, ... | | 著者 | Happonen, L.J, Oksanen, E, Kajander, T, Goldman, A, Butcher, S. | | 登録日 | 2013-04-27 | | 公開日 | 2013-05-22 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.892 Å) | | 主引用文献 | The Structure of the NTPase That Powers DNA Packaging into Sulfolobus Turreted Icosahedral Virus 2.
J.Virol., 87, 2013
|
|
6PSX
 
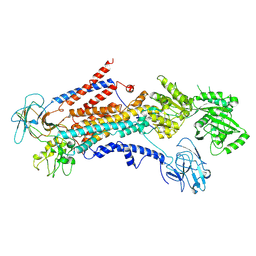 | | Cryo-EM structure of S. cerevisiae Drs2p-Cdc50p in the PI4P-activated form | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cell division control protein 50, ... | | 著者 | Bai, L, Li, H. | | 登録日 | 2019-07-14 | | 公開日 | 2019-09-25 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Autoinhibition and activation mechanisms of the eukaryotic lipid flippase Drs2p-Cdc50p.
Nat Commun, 10, 2019
|
|
7SOB
 
 | |
3MGP
 
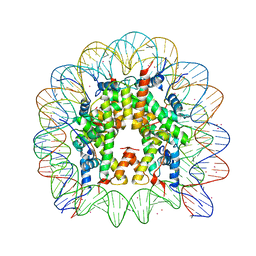 | | Binding of Cobalt ions to the Nucleosome Core Particle | | 分子名称: | CHLORIDE ION, COBALT (II) ION, DNA (147-MER), ... | | 著者 | Mohideen, K, Muhammad, R, Davey, C.A. | | 登録日 | 2010-04-07 | | 公開日 | 2010-06-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | Perturbations in nucleosome structure from heavy metal association.
Nucleic Acids Res., 38, 2010
|
|
3DAB
 
 | |
4KGC
 
 | | Nucleosome Core Particle Containing (ETA6-P-CYMENE)-(1, 2-ETHYLENEDIAMINE)-RUTHENIUM | | 分子名称: | (ethane-1,2-diamine-kappa~2~N,N')[(1,2,3,4,5,6-eta)-1-methyl-4-(propan-2-yl)cyclohexane-1,2,3,4,5,6-hexayl]ruthenium, DNA (145-mer), Histone H2A, ... | | 著者 | Adhireksan, Z, Davey, C.A. | | 登録日 | 2013-04-29 | | 公開日 | 2014-03-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Ligand substitutions between ruthenium-cymene compounds can control protein versus DNA targeting and anticancer activity
Nat Commun, 5, 2014
|
|
3DAQ
 
 | | Crystal structure of dihydrodipicolinate synthase from methicillin-resistant Staphylococcus aureus | | 分子名称: | CHLORIDE ION, Dihydrodipicolinate synthase, GLYCEROL | | 著者 | Dobson, R.C.J, Burgess, B.R, Jameson, G.B, Gerrard, J.A, Parker, M.W, Perugini, M.A. | | 登録日 | 2008-05-29 | | 公開日 | 2008-08-05 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structure and evolution of a novel dimeric enzyme from a clinically-important bacterial pathogen.
J.Biol.Chem., 2008
|
|
4KH6
 
 | |
