4XPC
 
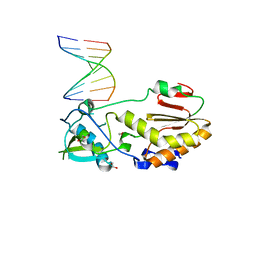 | |
4XPE
 
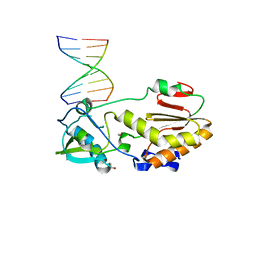 | |
4FMN
 
 | | Structure of the C-terminal domain of the Saccharomyces cerevisiae MUTL alpha (MLH1/PMS1) heterodimer bound to a fragment of NTG2 | | 分子名称: | 1,2-ETHANEDIOL, DNA mismatch repair protein MLH1, DNA mismatch repair protein PMS1, ... | | 著者 | Gueneau, E, Legrand, P, Charbonnier, J.B. | | 登録日 | 2012-06-18 | | 公開日 | 2013-02-20 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Structure of the MutL alpha C-terminal domain reveals how Mlh1 contributes to Pms1 endonuclease site.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4YM5
 
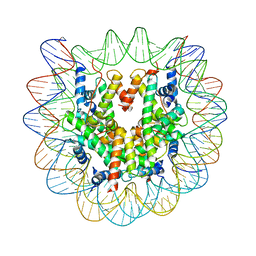 | | Crystal structure of the human nucleosome containing 6-4PP (inside) | | 分子名称: | 144 mer-DNA, 144-mer DNA, Histone H2A type 1-B/E, ... | | 著者 | Osakabe, A, Tachiwana, H, Kagawa, W, Horikoshi, N, Matsumoto, S, Hasegawa, M, Matsumoto, N, Toga, T, Yamamoto, J, Hanaoka, F, Thoma, N.H, Sugasawa, K, Iwai, S, Kurumizaka, H. | | 登録日 | 2015-03-06 | | 公開日 | 2015-12-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (4.005 Å) | | 主引用文献 | Structural basis of pyrimidine-pyrimidone (6-4) photoproduct recognition by UV-DDB in the nucleosome
Sci Rep, 5, 2015
|
|
2W57
 
 | |
7BM2
 
 | |
8VCJ
 
 | |
4GS3
 
 | | Dimeric structure of the N-terminal domain of PriB protein from Thermoanaerobacter tencongensis solved ab initio | | 分子名称: | Single-stranded DNA-binding protein | | 著者 | Liebschner, D, Brzezinski, K, Dauter, M, Dauter, Z, Nowak, M, Kur, J, Olszewski, M. | | 登録日 | 2012-08-27 | | 公開日 | 2012-09-19 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.09 Å) | | 主引用文献 | Dimeric structure of the N-terminal domain of PriB protein from Thermoanaerobacter tengcongensis solved ab initio.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5HOO
 
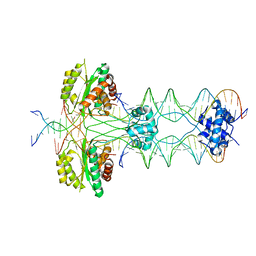 | |
7LMA
 
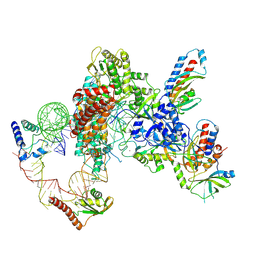 | | Tetrahymena telomerase T3D2 structure at 3.3 Angstrom | | 分子名称: | Telomerase La-related protein p65, Telomerase RNA, Telomerase associated protein p50, ... | | 著者 | He, Y, Wang, Y, Liu, B, Helmling, C, Susac, L, Cheng, R, Zhou, Z.H, Feigon, J. | | 登録日 | 2021-02-05 | | 公開日 | 2021-05-12 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structures of telomerase at several steps of telomere repeat synthesis.
Nature, 593, 2021
|
|
7LMB
 
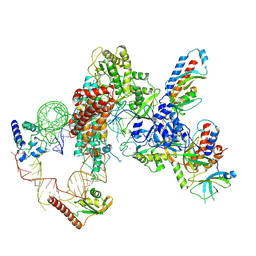 | | Tetrahymena telomerase T5D5 structure at 3.8 Angstrom | | 分子名称: | Telomerase La-related protein p65, Telomerase RNA, Telomerase associated protein p50, ... | | 著者 | He, Y, Wang, Y, Liu, B, Helmling, C, Susac, L, Cheng, R, Zhou, Z.H, Feigon, J. | | 登録日 | 2021-02-05 | | 公開日 | 2021-05-12 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structures of telomerase at several steps of telomere repeat synthesis.
Nature, 593, 2021
|
|
6ZX9
 
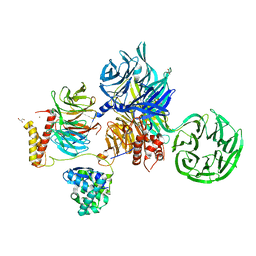 | | Crystal structure of SIV Vpr,fused to T4 lysozyme, isolated from moustached monkey, bound to human DDB1 and human DCAF1 (amino acid residues 1046-1396) | | 分子名称: | DDB1- and CUL4-associated factor 1, DNA damage-binding protein 1, GLYCEROL, ... | | 著者 | Schwefel, D, Banchenko, S. | | 登録日 | 2020-07-29 | | 公開日 | 2021-07-21 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.519729 Å) | | 主引用文献 | Structural insights into Cullin4-RING ubiquitin ligase remodelling by Vpr from simian immunodeficiency viruses.
Plos Pathog., 17, 2021
|
|
5IWM
 
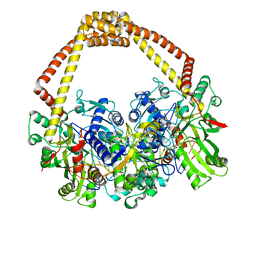 | | 2.5A structure of GSK945237 with S.aureus DNA gyrase and DNA. | | 分子名称: | (1R)-1-[(4-{[(6,7-dihydro[1,4]dioxino[2,3-c]pyridazin-3-yl)methyl]amino}piperidin-1-yl)methyl]-9-fluoro-1,2-dihydro-4H-pyrrolo[3,2,1-ij]quinolin-4-one, DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*GP*GP*TP*TP*CP*AP*CP*CP*GP*CP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*GP*CP*GP*GP*TP*GP*AP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), ... | | 著者 | Bax, B.D, Miles, T.J. | | 登録日 | 2016-03-22 | | 公開日 | 2016-05-25 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Novel tricyclics (e.g., GSK945237) as potent inhibitors of bacterial type IIA topoisomerases.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
8W0E
 
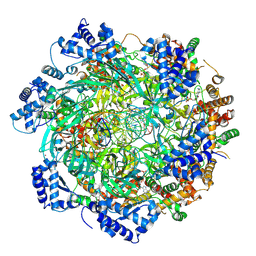 | |
8W0F
 
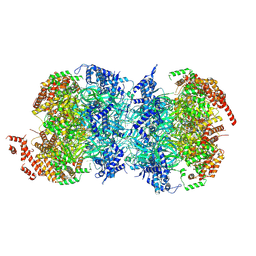 | |
5ZBA
 
 | | Crystal structure of Rtt109-Asf1-H3-H4-CoA complex | | 分子名称: | COENZYME A, DNA damage response protein Rtt109, putative, ... | | 著者 | Zhang, L, Serra-Cardona, A, Zhou, H, Wang, M, Yang, N, Zhang, Z, Xu, R.M. | | 登録日 | 2018-02-10 | | 公開日 | 2018-07-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Multisite Substrate Recognition in Asf1-Dependent Acetylation of Histone H3 K56 by Rtt109.
Cell, 174, 2018
|
|
1I6H
 
 | | RNA POLYMERASE II ELONGATION COMPLEX | | 分子名称: | 5'-D(P*AP*AP*AP*TP*GP*CP*CP*TP*GP*GP*TP*CP*T)-3', 5'-R(P*GP*AP*CP*CP*AP*GP*GP*CP*A)-3', DNA-DIRECTED RNA POLYMERASE II 13.6KD POLYPEPTIDE, ... | | 著者 | Gnatt, A.L, Cramer, P, Kornberg, R.D. | | 登録日 | 2001-03-02 | | 公開日 | 2001-04-23 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution.
Science, 292, 2001
|
|
5ZBB
 
 | | Crystal structure of Rtt109-Asf1-H3-H4 complex | | 分子名称: | DI(HYDROXYETHYL)ETHER, DNA damage response protein Rtt109, putative, ... | | 著者 | Zhang, L, Serra-Cardona, A, Zhou, H, Wang, M, Yang, N, Zhang, Z, Xu, R.M. | | 登録日 | 2018-02-10 | | 公開日 | 2018-07-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Multisite Substrate Recognition in Asf1-Dependent Acetylation of Histone H3 K56 by Rtt109.
Cell, 174, 2018
|
|
1PVR
 
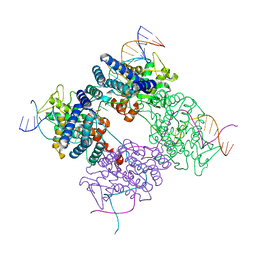 | | BASIS FOR A SWITCH IN SUBSTRATE SPECIFICITY: CRYSTAL STRUCTURE OF SELECTED VARIANT OF CRE SITE-SPECIFIC RECOMBINASE, LNSGG BOUND TO THE LOXP (WILDTYPE) RECOGNITION SITE | | 分子名称: | 34-MER, Recombinase CRE | | 著者 | Baldwin, E.P, Martin, S.S, Abel, J, Gelato, K.A, Kim, H, Schultz, P.G, Santoro, S.W. | | 登録日 | 2003-06-28 | | 公開日 | 2004-02-17 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | A specificity switch in selected cre recombinase variants is mediated by macromolecular plasticity and water.
Chem.Biol., 10, 2003
|
|
1PVP
 
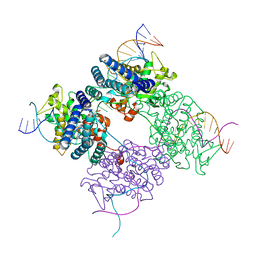 | | BASIS FOR A SWITCH IN SUBSTRATE SPECIFICITY: CRYSTAL STRUCTURE OF SELECTED VARIANT OF CRE SITE-SPECIFIC RECOMBINASE, ALSHG BOUND TO THE ENGINEERED RECOGNITION SITE LOXM7 | | 分子名称: | 34-MER, Recombinase cre | | 著者 | Baldwin, E.P, Martin, S.S, Abel, J, Gelato, K.A, Kim, H, Schultz, P.G, Santoro, S.W. | | 登録日 | 2003-06-28 | | 公開日 | 2004-02-17 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | A specificity switch in selected cre recombinase variants is mediated by macromolecular plasticity and water.
Chem.Biol., 10, 2003
|
|
1MYK
 
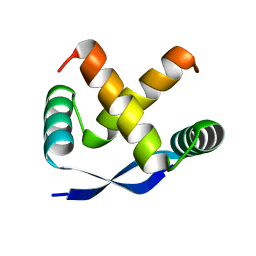 | | CRYSTAL STRUCTURE, FOLDING, AND OPERATOR BINDING OF THE HYPERSTABLE ARC REPRESSOR MUTANT PL8 | | 分子名称: | ARC REPRESSOR | | 著者 | Schildbach, J.F, Milla, M.E, Jeffrey, P.D, Raumann, B.E, Sauer, R.T. | | 登録日 | 1994-10-12 | | 公開日 | 1995-01-26 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure, folding, and operator binding of the hyperstable Arc repressor mutant PL8.
Biochemistry, 34, 1995
|
|
2BJ8
 
 | |
1EYF
 
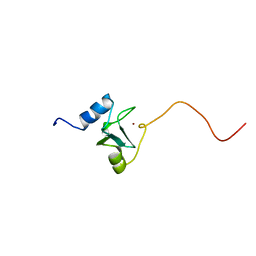 | | REFINED STRUCTURE OF THE DNA METHYL PHOSPHOTRIESTER REPAIR DOMAIN OF E. COLI ADA | | 分子名称: | ADA REGULATORY PROTEIN, ZINC ION | | 著者 | Lin, Y, Dotsch, V, Wintner, T, Peariso, K, Myers, L.C, Penner-Hahn, J.E, Verdine, G.L, Wagner, G. | | 登録日 | 2000-05-06 | | 公開日 | 2003-09-09 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for the functional switch of the E. coli Ada protein
Biochemistry, 40, 2001
|
|
6URS
 
 | | Sleeping Beauty transposase PAI subdomain mutant - H19Y | | 分子名称: | Sleeping Beauty transposase PAI subdomain | | 著者 | Nesmelova, I.V, Leighton, G.O, Yan, C, Lustig, J, Corona, R.I, Guo, J.T, Ivics, Z. | | 登録日 | 2019-10-24 | | 公開日 | 2020-10-28 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | H19Y mutation in the primary DNA-recognition subdomain of the Sleeping Beauty transposase improves structural stability, transposon DNA-binding and transposition
To Be Published
|
|
2BJ7
 
 | |
