7B3B
 
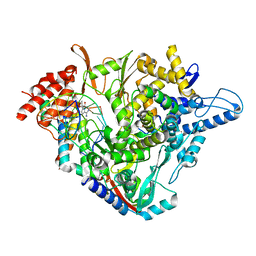 | | Structure of elongating SARS-CoV-2 RNA-dependent RNA polymerase with Remdesivir at position -3 (structure 1) | | 分子名称: | DNA/RNA (5'-R(P*CP*UP*AP*CP*GP*CP*G)-D(P*(RMP))-R(P*UP*G)-3'), Non-structural protein 7, Non-structural protein 8, ... | | 著者 | Kokic, G, Hillen, H.S, Tegunov, D, Dienemann, C, Seitz, F, Schmitzova, J, Farnung, L, Siewert, A, Hoebartner, C, Cramer, P. | | 登録日 | 2020-11-30 | | 公開日 | 2020-12-23 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Mechanism of SARS-CoV-2 polymerase stalling by remdesivir.
Nat Commun, 12, 2021
|
|
7KVL
 
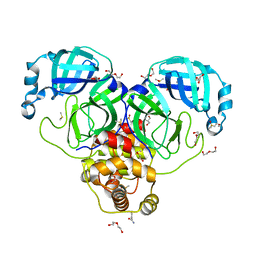 | | SARS-CoV-2 Main protease immature form - FMAX Library E01 fragment | | 分子名称: | 2-chloropyridine-4-carboxamide, 3C-like proteinase, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Noske, G.D, Nakamura, A.M, Gawriljuk, V.O, Lima, G.M.A, Zeri, A.C.M, Nascimento, A.F.Z, Fernandes, R.S, Oliva, G, Godoy, A.S. | | 登録日 | 2020-11-28 | | 公開日 | 2020-12-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | A Crystallographic Snapshot of SARS-CoV-2 Main Protease Maturation Process.
J.Mol.Biol., 433, 2021
|
|
7KVR
 
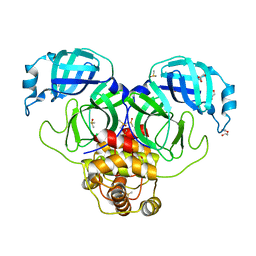 | | SARS-CoV-2 Main protease immature form - FMAX Library E09 fragment | | 分子名称: | 3C-like proteinase, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | 著者 | Noske, G.D, Nakamura, A.M, Gawriljuk, V.O, Lima, G.M.A, Zeri, A.C.M, Nascimento, A.F.Z, Fernandes, R.S, Oliva, G, Godoy, A.S. | | 登録日 | 2020-11-28 | | 公開日 | 2020-12-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | A Crystallographic Snapshot of SARS-CoV-2 Main Protease Maturation Process.
J.Mol.Biol., 433, 2021
|
|
7B2U
 
 | |
7B2J
 
 | |
7B27
 
 | |
7DK4
 
 | |
7DK7
 
 | |
7DK5
 
 | |
7DK6
 
 | |
7DK3
 
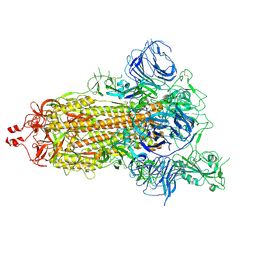 | | SARS-CoV-2 S trimer, S-open | | 分子名称: | Spike glycoprotein | | 著者 | Xu, C, Cong, Y. | | 登録日 | 2020-11-23 | | 公開日 | 2020-12-16 | | 最終更新日 | 2021-02-17 | | 実験手法 | ELECTRON MICROSCOPY (6 Å) | | 主引用文献 | Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM.
Sci Adv, 7, 2021
|
|
7DK1
 
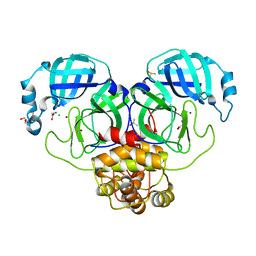 | | Crystal structure of Zinc bound SARS-CoV-2 main protease | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3C-like proteinase, CHLORIDE ION, ... | | 著者 | Sonkar, K.S, Panchariya, L, Kuila, S, Khan, W.A, Arockiasamy, A. | | 登録日 | 2020-11-22 | | 公開日 | 2021-06-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.902 Å) | | 主引用文献 | Zinc 2+ ion inhibits SARS-CoV-2 main protease and viral replication in vitro.
Chem.Commun.(Camb.), 57, 2021
|
|
7KS9
 
 | |
7DJR
 
 | |
7KRX
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder441 inhibitor | | 分子名称: | 3-amino-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, ACETATE ION, CHLORIDE ION, ... | | 著者 | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-11-20 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.72 Å) | | 主引用文献 | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder441
to be published
|
|
7DHX
 
 | | Crystal structure of SARS-CoV-2 RBD binding to pangolin ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ZINC ION, ... | | 著者 | Wang, Q.H, Qi, J.X, Wu, L.L. | | 登録日 | 2020-11-17 | | 公開日 | 2021-09-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Molecular basis of pangolin ACE2 engaged by COVID-19 virus
Chin.Sci.Bull., 66, 2021
|
|
7DHJ
 
 | | The co-crystal structure of SARS-CoV-2 main protease with the peptidomimetic inhibitor (S)-2-cinnamamido-N-((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)pent-4-ynamide | | 分子名称: | (2~{S})-~{N}-[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-2-[[(~{E})-3-phenylprop-2-enoyl]amino]pent-4-ynamide, 3C-like proteinase | | 著者 | Shang, L.Q, Wang, H, Deng, W.L, Xing, S, Wang, Y.X. | | 登録日 | 2020-11-15 | | 公開日 | 2021-11-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.962 Å) | | 主引用文献 | The structure-based design of peptidomimetic inhibitors against SARS-CoV-2 3C like protease as Potent anti-viral drug candidate.
Eur.J.Med.Chem., 238, 2022
|
|
7KQE
 
 | |
7KQB
 
 | | SARS-CoV-2 spike glycoprotein:Fab 5A6 complex I | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 5A6 heavy chain, Fab 5A6 light chain, ... | | 著者 | Asarnow, D, Charles, C, Cheng, Y. | | 登録日 | 2020-11-14 | | 公開日 | 2021-05-26 | | 最終更新日 | 2021-06-23 | | 実験手法 | ELECTRON MICROSCOPY (2.42 Å) | | 主引用文献 | Structural insight into SARS-CoV-2 neutralizing antibodies and modulation of syncytia.
Cell, 184, 2021
|
|
5S72
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with BBL029427 | | 分子名称: | CITRIC ACID, N-(2-aminoethyl)-N'-phenylurea, Uridylate-specific endoribonuclease | | 著者 | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | 登録日 | 2020-11-13 | | 公開日 | 2020-11-25 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.512 Å) | | 主引用文献 | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5S6Y
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z56900771 | | 分子名称: | CITRIC ACID, N-[(furan-2-yl)methyl]urea, Uridylate-specific endoribonuclease | | 著者 | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | 登録日 | 2020-11-13 | | 公開日 | 2020-11-25 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5S6Z
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with PB2255187532 | | 分子名称: | 4-[(dimethylamino)methyl]-1,3-thiazol-2-amine, CITRIC ACID, Uridylate-specific endoribonuclease | | 著者 | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | 登録日 | 2020-11-13 | | 公開日 | 2020-11-25 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5S71
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with FUZS-5 | | 分子名称: | 5'-thiothymidine, CITRIC ACID, Uridylate-specific endoribonuclease | | 著者 | Godoy, A.S, Bege, M, Bajusz, D, BorbAs, A, Keseru, G.M, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | 登録日 | 2020-11-13 | | 公開日 | 2020-11-25 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.941 Å) | | 主引用文献 | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5S6X
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z2889976755 | | 分子名称: | 1-(2,4-dimethyl-1H-imidazol-5-yl)methanamine, CITRIC ACID, Uridylate-specific endoribonuclease | | 著者 | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | 登録日 | 2020-11-13 | | 公開日 | 2020-11-25 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5S70
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with EN300-181428 | | 分子名称: | (5R)-2-methyl-4,5,6,7-tetrahydro-1H-benzimidazol-5-amine, CITRIC ACID, Uridylate-specific endoribonuclease | | 著者 | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | 登録日 | 2020-11-13 | | 公開日 | 2020-11-25 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.327 Å) | | 主引用文献 | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
