3MNR
 
 | | Crystal Structure of Benzamide SNX-1321 bound to Hsp90 | | 分子名称: | 2-[(3,4,5-trimethoxyphenyl)amino]-4-(2,6,6-trimethyl-4-oxo-4,5,6,7-tetrahydro-1H-indol-1-yl)benzamide, Heat shock protein HSP 90-alpha | | 著者 | Veal, J.M, Fadden, P, Huang, K.H, Rice, J, Hall, S.E, Haytstead, T.A. | | 登録日 | 2010-04-22 | | 公開日 | 2010-08-11 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Application of Chemoproteomics to Drug Discovery: Identification of a Clinical Candidate Targeting Hsp90.
Chem.Biol., 17, 2010
|
|
3MH5
 
 | | HtrA proteases are activated by a conserved mechanism that can be triggered by distinct molecular cues | | 分子名称: | DIISOPROPYL PHOSPHONATE, Protease do | | 著者 | Krojer, T, Sawa, J, Huber, R, Clausen, T. | | 登録日 | 2010-04-07 | | 公開日 | 2010-06-30 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | HtrA proteases have a conserved activation mechanism that can be triggered by distinct molecular cues
Nat.Struct.Mol.Biol., 17, 2010
|
|
3MOJ
 
 | |
6QPZ
 
 | | Crystal structure of as isolated Y323E mutant of haem-Cu containing nitrite reductase from Ralstonia pickettii | | 分子名称: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | 著者 | Antonyuk, S.V, Shenoy, R.T, Hedison, T.M, Eady, R.R, Hasnain, S.S, Scrutton, N.S. | | 登録日 | 2019-02-16 | | 公開日 | 2019-11-06 | | 最終更新日 | 2020-02-26 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Unexpected Roles of a Tether Harboring a Tyrosine Gatekeeper Residue in Modular Nitrite Reductase Catalysis.
Acs Catalysis, 9, 2019
|
|
3MJ8
 
 | |
3MQH
 
 | | crystal structure of the 3-N-acetyl transferase WlbB from Bordetella petrii in complex with CoA and UDP-3-amino-2-acetamido-2,3-dideoxy glucuronic acid | | 分子名称: | (2S,3S,4R,5R,6R)-5-(acetylamino)-4-amino-6-{[(R)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-3-hydroxytetrahydro-2H-pyran-2-carboxylic acid, 1,2-ETHANEDIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ... | | 著者 | thoden, J.B, holden, H.M. | | 登録日 | 2010-04-28 | | 公開日 | 2010-05-12 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | Molecular structure of WlbB, a bacterial N-acetyltransferase involved in the biosynthesis of 2,3-diacetamido-2,3-dideoxy-D-mannuronic acid .
Biochemistry, 49, 2010
|
|
3MJS
 
 | | Structure of A-type Ketoreductases from Modular Polyketide Synthase | | 分子名称: | (2S)-2-hydroxybutanedioic acid, AmphB, D-MALATE, ... | | 著者 | Zheng, J, Taylor, C.A, Piasecki, S.K, Keatinge-Clay, A.T. | | 登録日 | 2010-04-13 | | 公開日 | 2010-08-18 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural and Functional Analysis of A-Type Ketoreductases from the Amphotericin Modular Polyketide Synthase.
Structure, 18, 2010
|
|
6QQC
 
 | |
3MKD
 
 | | Crystal structure of myosin-2 dictyostelium discoideum motor domain S456Y mutant in complex with adp-orthovanadate | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Myosin-2 heavy chain, ... | | 著者 | Kathmann, D, Diensthuber, R.P, Fedorov, R, Manstein, D.J, Tsiavaliaris, G. | | 登録日 | 2010-04-14 | | 公開日 | 2011-04-06 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Switch-2 dependent modulation of the myosin power stroke
To be Published
|
|
3MKW
 
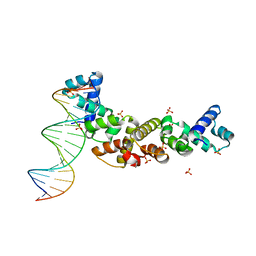 | | Structure of sopB(155-272)-18mer complex, I23 form | | 分子名称: | DNA (5'-D(*CP*TP*GP*GP*GP*AP*CP*CP*AP*TP*GP*GP*TP*CP*CP*CP*AP*G)-3'), Protein sopB, SULFATE ION | | 著者 | Schumacher, M.A, Piro, K, Xu, W. | | 登録日 | 2010-04-15 | | 公開日 | 2010-05-05 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.99 Å) | | 主引用文献 | Insight into F plasmid DNA segregation revealed by structures of SopB and SopB-DNA complexes.
Nucleic Acids Res., 38, 2010
|
|
6DB4
 
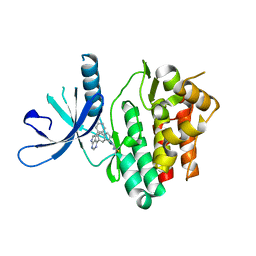 | | JAK3 with Cyanamide CP34 | | 分子名称: | N-[(1S)-6-(5-phenyl-7H-pyrrolo[2,3-d]pyrimidin-4-yl)-2,3-dihydro-1H-inden-1-yl]imidoformamide, Tyrosine-protein kinase JAK3 | | 著者 | Vajdos, F.F. | | 登録日 | 2018-05-02 | | 公開日 | 2018-11-28 | | 最終更新日 | 2019-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.662 Å) | | 主引用文献 | Identification of Cyanamide-Based Janus Kinase 3 (JAK3) Covalent Inhibitors.
J. Med. Chem., 61, 2018
|
|
6DC3
 
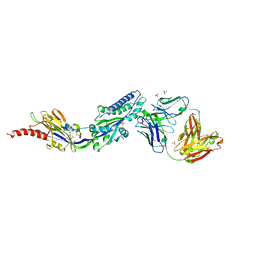 | | RSV prefusion F bound to RSD5 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab RSD5-Germline Heavy Chain, Fab RSD5-Germline Light Chain, ... | | 著者 | Battles, M.B, McLellan, J.S, Jones, H.J. | | 登録日 | 2018-05-04 | | 公開日 | 2019-07-10 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.501 Å) | | 主引用文献 | Alternative conformations of a major antigenic site on RSV F.
Plos Pathog., 15, 2019
|
|
3MMK
 
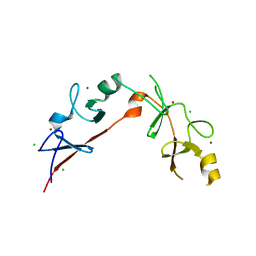 | | The structural basis for partial redundancy in a class of transcription factors, the lim-homeodomain proteins, in neural cell type specification | | 分子名称: | CHLORIDE ION, Fusion of LIM/homeobox protein Lhx4, linker, ... | | 著者 | Gadd, M.S, Langley, D.B, Guss, J.M, Matthews, J.M. | | 登録日 | 2010-04-20 | | 公開日 | 2011-07-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.157 Å) | | 主引用文献 | The structural basis for partial redundancy in a class of transcription factors, the lim-homeodomain proteins, in neural cell type specification.
J.Biol.Chem., 2011
|
|
3M3T
 
 | |
3MP4
 
 | | Crystal structure of Human lyase R41M mutant | | 分子名称: | Hydroxymethylglutaryl-CoA lyase | | 著者 | Fu, Z, Runquist, J.A, Montgomery, C, Miziorko, H.M, Kim, J.-J.P. | | 登録日 | 2010-04-24 | | 公開日 | 2010-06-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Functional insights into human HMG-CoA lyase from structures of Acyl-CoA-containing ternary complexes.
J.Biol.Chem., 285, 2010
|
|
3M3Z
 
 | | Crystal structure of HSC70/BAG1 in complex with small molecule inhibitor | | 分子名称: | 5'-O-(2-amino-2-oxoethyl)-8-(methylamino)adenosine, BAG family molecular chaperone regulator 1, Heat shock cognate 71 kDa protein | | 著者 | Dokurno, P, Surgenor, A.E, Shaw, T, Macias, A.T, Massey, A.J, Williamson, D.S. | | 登録日 | 2010-03-10 | | 公開日 | 2011-01-26 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Adenosine-Derived Inhibitors of 78 kDa Glucose Regulated Protein (Grp78) ATPase: Insights into Isoform Selectivity.
J.Med.Chem., 54, 2011
|
|
6QTX
 
 | |
6QUM
 
 | |
6DU5
 
 | |
3MSL
 
 | |
6QSZ
 
 | | Crystal structure of the Sir4 H-BRCT domain in complex with Esc1 pS1450 peptide | | 分子名称: | CHLORIDE ION, Regulatory protein SIR4, Silent chromatin protein ESC1 | | 著者 | Deshpande, I, Keusch, J.J, Challa, K, Iesmantavicius, V, Gasser, S.M, Gut, H. | | 登録日 | 2019-02-22 | | 公開日 | 2019-09-18 | | 最終更新日 | 2019-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The Sir4 H-BRCT domain interacts with phospho-proteins to sequester and repress yeast heterochromatin.
Embo J., 38, 2019
|
|
3M0B
 
 | | Ru-Porphyrin Protein Scaffolds for Sensing O2 | | 分子名称: | CARBON MONOXIDE, Methyl-accepting chemotaxis protein, [3,3'-(7,12-diethyl-3,8,13,17-tetramethylporphyrin-2,18-diyl-kappa~4~N~21~,N~22~,N~23~,N~24~)dipropanoato(2-)]ruthenium | | 著者 | Winter, M.B, McLaurin, E.J, Reece, S.Y, Olea Jr, C, Nocera, D.G, Marletta, M.A. | | 登録日 | 2010-03-02 | | 公開日 | 2010-04-14 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Ru-porphyrin protein scaffolds for sensing O2.
J.Am.Chem.Soc., 132, 2010
|
|
3MEC
 
 | | HIV-1 Reverse Transcriptase in Complex with TMC125 | | 分子名称: | 4-({6-AMINO-5-BROMO-2-[(4-CYANOPHENYL)AMINO]PYRIMIDIN-4-YL}OXY)-3,5-DIMETHYLBENZONITRILE, SULFATE ION, p51 Reverse transcriptase, ... | | 著者 | Lansdon, E.B. | | 登録日 | 2010-03-31 | | 公開日 | 2010-05-12 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal Structures of HIV-1 Reverse Transcriptase with Etravirine (TMC125) and Rilpivirine (TMC278): Implications for Drug Design.
J.Med.Chem., 53, 2010
|
|
6DPS
 
 | |
3MA2
 
 | | Complex membrane type-1 matrix metalloproteinase (MT1-MMP) with tissue inhibitor of metalloproteinase-1 (TIMP-1) | | 分子名称: | CALCIUM ION, Matrix metalloproteinase-14, Metalloproteinase inhibitor 1, ... | | 著者 | Grossman, M, Tworowski, D, Dym, O, Lee, M.-H, Levy, Y, Sagi, I. | | 登録日 | 2010-03-23 | | 公開日 | 2010-06-30 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | The Intrinsic Protein Flexibility of Endogenous Protease Inhibitor TIMP-1 Controls Its Binding Interface and Affects Its Function.
Biochemistry, 49, 2010
|
|
