2LXY
 
 | | NMR structure of 2-MERCAPTOPHENOL-ALPHA3C | | 分子名称: | 2-MERCAPTOPHENOL, 2-mercaptophenol-alpha3C | | 著者 | Tommos, C, Valentine, K.G, Martinez-Rivera, M.C, Liang, L, Moorman, V.R. | | 登録日 | 2012-09-06 | | 公開日 | 2013-02-27 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Reversible phenol oxidation and reduction in the structurally well-defined 2-Mercaptophenol-alpha(3)C protein.
Biochemistry, 52, 2013
|
|
4YY2
 
 | |
3BAR
 
 | | Crystal structure of Plasmodium falciparum orotidine 5'-phosphate decarboxylase covalently modified by 6-azido-UMP | | 分子名称: | Orotidine 5'-phosphate decarboxylase, URIDINE-5'-MONOPHOSPHATE | | 著者 | Liu, Y, Bello, A.M, Poduch, E, Lau, W, Kotra, L.P, Pai, E.F. | | 登録日 | 2007-11-08 | | 公開日 | 2008-01-29 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure-Activity Relationships of C6-Uridine Derivatives Targeting Plasmodia Orotidine Monophosphate Decarboxylase.
J.Med.Chem., 51, 2008
|
|
1BWS
 
 | | CRYSTAL STRUCTURE OF GDP-4-KETO-6-DEOXY-D-MANNOSE EPIMERASE/REDUCTASE FROM ESCHERICHIA COLI A KEY ENZYME IN THE BIOSYNTHESIS OF GDP-L-FUCOSE | | 分子名称: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTEIN (GDP-4-KETO-6-DEOXY-D-MANNOSE EPIMERASE/REDUCTASE) | | 著者 | Rizzi, M, Tonetti, M, Flora, A.D, Bolognesi, M. | | 登録日 | 1998-09-25 | | 公開日 | 1999-01-13 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | GDP-4-keto-6-deoxy-D-mannose epimerase/reductase from Escherichia coli, a key enzyme in the biosynthesis of GDP-L-fucose, displays the structural characteristics of the RED protein homology superfamily.
Structure, 6, 1998
|
|
4YXY
 
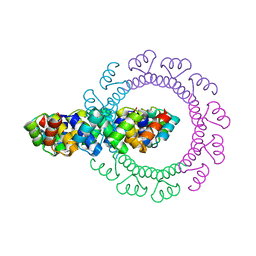 | |
1CDE
 
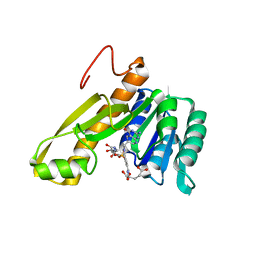 | | STRUCTURES OF APO AND COMPLEXED ESCHERICHIA COLI GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE | | 分子名称: | 5-DEAZAFOLIC ACID, GLYCINAMIDE RIBONUCLEOTIDE, PHOSPHORIBOSYL-GLYCINAMIDE FORMYLTRANSFERASE | | 著者 | Almassy, R.J, Janson, C.A, Kan, C.-C, Hostomska, Z. | | 登録日 | 1992-05-15 | | 公開日 | 1993-10-31 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structures of apo and complexed Escherichia coli glycinamide ribonucleotide transformylase.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
4YXX
 
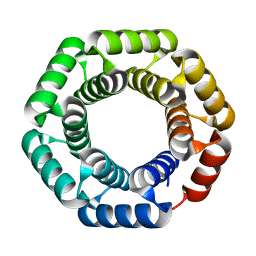 | |
4YXZ
 
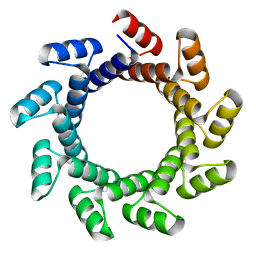 | |
4YY5
 
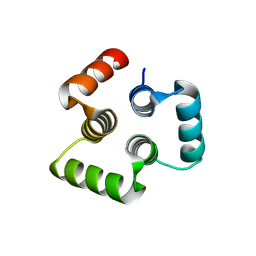 | |
1DOF
 
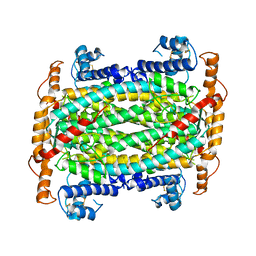 | | THE CRYSTAL STRUCTURE OF ADENYLOSUCCINATE LYASE FROM PYROBACULUM AEROPHILUM: INSIGHTS INTO THERMAL STABILITY AND HUMAN PATHOLOGY | | 分子名称: | ADENYLOSUCCINATE LYASE | | 著者 | Toth, E.A, Yeates, T.O, Goedken, E, Dixon, J.E, Marqusee, S. | | 登録日 | 1999-12-20 | | 公開日 | 2001-01-10 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The crystal structure of adenylosuccinate lyase from Pyrobaculum aerophilum reveals an intracellular protein with three disulfide bonds.
J.Mol.Biol., 301, 2000
|
|
5BYO
 
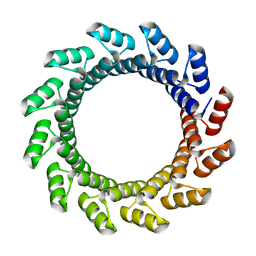 | |
3DGO
 
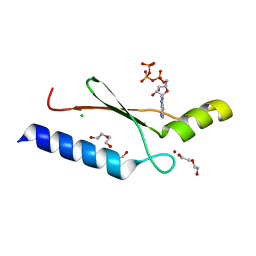 | | A non-biological ATP binding protein with a Tyr-Phe mutation in the ligand binding domain | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, ATP Binding Protein-DX, CHLORIDE ION, ... | | 著者 | Simmons, C.R, Allen, J.P, Chaput, J.C. | | 登録日 | 2008-06-13 | | 公開日 | 2009-06-30 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A synthetic protein selected for ligand binding affinity mediates ATP hydrolysis.
Acs Chem.Biol., 4, 2009
|
|
2GJF
 
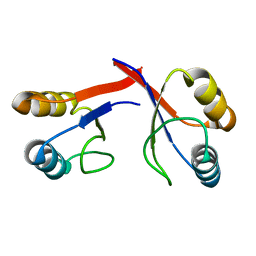 | |
3DGN
 
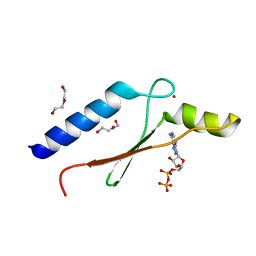 | | A non-biological ATP binding protein crystallized in the presence of 100 mM ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP Binding Protein-DX, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Simmons, C.R, Allen, J.P, Chaput, J.C. | | 登録日 | 2008-06-13 | | 公開日 | 2009-06-30 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | A synthetic protein selected for ligand binding affinity mediates ATP hydrolysis.
Acs Chem.Biol., 4, 2009
|
|
3DGL
 
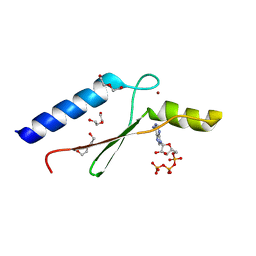 | | 1.8 A Crystal Structure of a Non-biological Protein with Bound ATP in a Novel Bent Conformation | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, ATP Binding Protein-DX, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Simmons, C.R, Allen, J.P, Chaput, J.C. | | 登録日 | 2008-06-13 | | 公開日 | 2009-06-30 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | A synthetic protein selected for ligand binding affinity mediates ATP hydrolysis.
Acs Chem.Biol., 4, 2009
|
|
1CDD
 
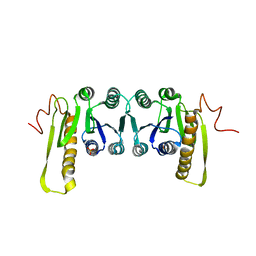 | | STRUCTURES OF APO AND COMPLEXED ESCHERICHIA COLI GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE | | 分子名称: | PHOSPHATE ION, PHOSPHORIBOSYL-GLYCINAMIDE FORMYLTRANSFERASE | | 著者 | Almassy, R.J, Janson, C.A, Kan, C.-C, Hostomska, Z. | | 登録日 | 1992-05-15 | | 公開日 | 1993-10-31 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structures of apo and complexed Escherichia coli glycinamide ribonucleotide transformylase.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
1EIX
 
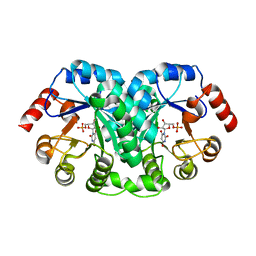 | | STRUCTURE OF OROTIDINE 5'-MONOPHOSPHATE DECARBOXYLASE FROM E. COLI, CO-CRYSTALLISED WITH THE INHIBITOR BMP | | 分子名称: | 1-(5'-PHOSPHO-BETA-D-RIBOFURANOSYL)BARBITURIC ACID, OROTIDINE 5'-MONOPHOSPHATE DECARBOXYLASE | | 著者 | Harris, P, Poulsen, J.C.N, Jensen, K.F, Larsen, S. | | 登録日 | 2000-02-29 | | 公開日 | 2000-03-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural basis for the catalytic mechanism of a proficient enzyme: orotidine 5'-monophosphate decarboxylase.
Biochemistry, 39, 2000
|
|
7FAO
 
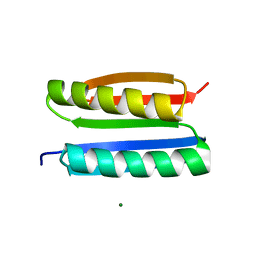 | |
2QAF
 
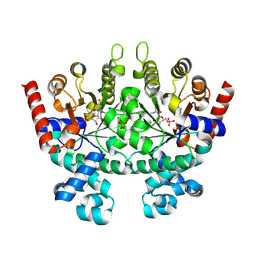 | | Crystal structure of Plasmodium falciparum orotidine 5'-phosphate decarboxylase covalently modified by 6-iodo-UMP | | 分子名称: | Orotidine 5' monophosphate decarboxylase, SULFATE ION, URIDINE-5'-MONOPHOSPHATE | | 著者 | Liu, Y, Lau, W, Bello, A.M, Kotra, L.P, Hui, R, Pai, E.F. | | 登録日 | 2007-06-15 | | 公開日 | 2008-04-29 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structure-activity relationships of C6-uridine derivatives targeting plasmodia orotidine monophosphate decarboxylase
J.Med.Chem., 51, 2008
|
|
4B02
 
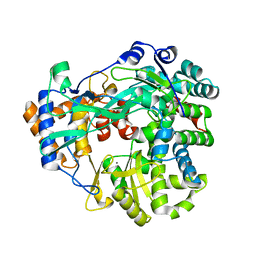 | | The C-terminal Priming Domain is Strongly Associated with the Main Body of Bacteriophage phi6 RNA-Dependent RNA Polymerase | | 分子名称: | MANGANESE (II) ION, RNA-DIRECTED RNA POLYMERASE | | 著者 | Sarin, L.P, Wright, S, Chen, Q, Degerth, L.H, Stuart, D.I, Grimes, J.M, Bamford, D.H, Poranen, M.M. | | 登録日 | 2012-06-27 | | 公開日 | 2012-08-01 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | The C-Terminal Priming Domain is Strongly Associated with the Main Body of Bacteriophage Phi6 RNA-Dependent RNA Polymerase.
Virology, 432, 2012
|
|
3ZWT
 
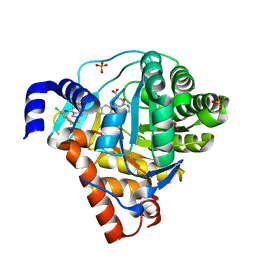 | | Structure of Human Dihydroorotate Dehydrogenase with a Bound Inhibitor | | 分子名称: | 2-{[(2,5-DICHLOROPHENYL)METHYL]SULFANYL}-5-ETHYL-[1,2,4]TRIAZOLO[1,5-A]PYRIMIDIN-7-OL, ACETATE ION, CHLORIDE ION, ... | | 著者 | Acklam, P.A, Parsons, M.R. | | 登録日 | 2011-08-02 | | 公開日 | 2012-06-27 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Factors Influencing the Specificity of Inhibitor Binding to the Human and Malaria Parasite Dihydroorotate Dehydrogenases.
J.Med.Chem., 55, 2012
|
|
3ZWS
 
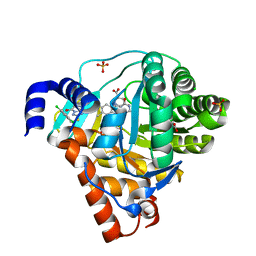 | | Structure of Human Dihydroorotate Dehydrogenase with a Bound Inhibitor | | 分子名称: | 2-[(2,5-DICHLOROBENZYL)SULFANYL]-5-METHYL[1,2,4]TRIAZOLO[1,5-A]PYRIMIDIN-7-OL, ACETATE ION, CHLORIDE ION, ... | | 著者 | Acklam, P.A, Parsons, M.R. | | 登録日 | 2011-08-02 | | 公開日 | 2012-06-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Factors Influencing the Specificity of Inhibitor Binding to the Human and Malaria Parasite Dihydroorotate Dehydrogenases.
J.Med.Chem., 55, 2012
|
|
7BQQ
 
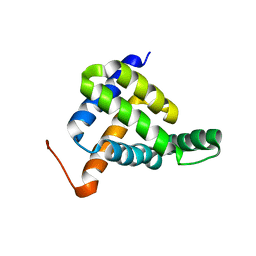 | | Solution NMR structure of fold-Z Gogy; de novo designed protein with an asymmetric all-alpha topology | | 分子名称: | Gogy | | 著者 | Kobayashi, N, Sugiki, T, Fujiwara, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | 登録日 | 2020-03-25 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
7BQM
 
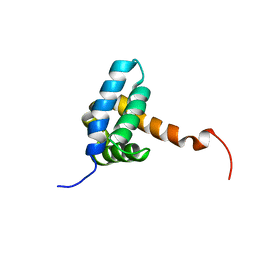 | | Solution NMR structure of fold-0 Chantal; de novo designed protein with an asymmetric all-alpha topology | | 分子名称: | Chantal | | 著者 | Kobayashi, N, Sugiki, T, Fujiwara, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | 登録日 | 2020-03-25 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
7BQN
 
 | | Solution NMR structure of fold-C Rei; de novo designed protein with an asymmetric all-alpha topology | | 分子名称: | Rei | | 著者 | Kobayashi, N, Sugiki, T, Fujiwara, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | 登録日 | 2020-03-25 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
