7BH8
 
 | | 3H4-Fab HLA-E-VL9 co-complex | | 分子名称: | 3H4 Fab heavy chain, 3H4 Fab light chain, Beta-2-microglobulin, ... | | 著者 | Walters, L.C, Rozbesky, D. | | 登録日 | 2021-01-10 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Mouse and human antibodies bind HLA-E-leader peptide complexes and enhance NK cell cytotoxicity.
Commun Biol, 5, 2022
|
|
8YYQ
 
 | | Structure of the HitB F328L mutant | | 分子名称: | Putative ATP-dependent b-aminoacyl-ACP synthetase, [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl ~{N}-[(3~{S})-3-azanyl-3-(3-cyanophenyl)propanoyl]sulfamate | | 著者 | Wang, D, Miyanaga, A, Chisuga, T, Kudo, F, Eguchi, T. | | 登録日 | 2024-04-04 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-08-14 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Engineering the Substrate Specificity of (S)-beta-Phenylalanine Adenylation Enzyme HitB.
Chembiochem, 25, 2024
|
|
4V1K
 
 | | SeMet structure of a novel carbohydrate binding module from glycoside hydrolase family 9 (Cel9A) from Ruminococcus flavefaciens FD-1 | | 分子名称: | 2-HYDROXY BUTANE-1,4-DIOL, CALCIUM ION, CARBOHYDRATE BINDING MODULE, ... | | 著者 | Venditto, I, Goyal, A, Thompson, A, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | 登録日 | 2014-09-29 | | 公開日 | 2016-01-20 | | 最終更新日 | 2018-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Complexity of the Ruminococcus Flavefaciens Cellulosome Reflects an Expansion in Glycan Recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7BCA
 
 | |
8Z28
 
 | |
4KI9
 
 | | Crystal structure of the catalytic domain of human DUSP12 at 2.0 A resolution | | 分子名称: | Dual specificity protein phosphatase 12, PHOSPHATE ION | | 著者 | Jeon, T.J, Chien, P.N, Ku, B, Kim, S.J, Ryu, S.E. | | 登録日 | 2013-05-02 | | 公開日 | 2014-02-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The family-wide structure and function of human dual-specificity protein phosphatases
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8YYR
 
 | | Structure of the HitB T293G mutant | | 分子名称: | Putative ATP-dependent b-aminoacyl-ACP synthetase, [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl ~{N}-[(3~{S})-3-azanyl-3-(2-bromophenyl)propanoyl]sulfamate | | 著者 | Wang, D, Miyanaga, A, Chisuga, T, Kudo, F, Eguchi, T. | | 登録日 | 2024-04-04 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-08-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Engineering the Substrate Specificity of (S)-beta-Phenylalanine Adenylation Enzyme HitB.
Chembiochem, 25, 2024
|
|
4KIS
 
 | | Crystal Structure of a LSR-DNA Complex | | 分子名称: | CALCIUM ION, DNA (26-MER), Putative integrase [Bacteriophage A118], ... | | 著者 | Rutherford, K, Yuan, P, Perry, K, Van Duyne, G.D. | | 登録日 | 2013-05-02 | | 公開日 | 2013-07-10 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Attachment site recognition and regulation of directionality by the serine integrases.
Nucleic Acids Res., 41, 2013
|
|
8XUP
 
 | |
9IK2
 
 | | The co-crystal structure of SARS-CoV-2 Mpro in complex with compound H109 | | 分子名称: | 3C-like proteinase, tert-butyl N-[(2S)-1-[[(2S)-1-[[(2S)-1-azanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-1-oxidanylidene-3-phenyl-propan-2-yl]amino]-3,3-dimethyl-1-oxidanylidene-butan-2-yl]carbamate | | 著者 | Feng, Y, Zheng, W.Y, Han, P, Fu, L.F, Qi, J.X. | | 登録日 | 2024-06-26 | | 公開日 | 2024-07-31 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure-guided discovery of a small molecule inhibitor of SARS-CoV-2 main protease with potent in vitro and in vivo antiviral activities
To Be Published
|
|
9CJA
 
 | |
8ZN3
 
 | | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter sp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.41 A resolution. | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, PHOSPHONOACETIC ACID, ... | | 著者 | Ahmad, N, Sharma, P, Bhushan, A, Sharma, S, Singh, T.P. | | 登録日 | 2024-05-25 | | 公開日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter sp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.41 A resolution.
To Be Published
|
|
7BJQ
 
 | |
6X0L
 
 | |
8XMT
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron EG.5.1 spike protein(6P), RBD-closed state | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Li, L.J, Gu, Y.H, Shi, K.Y, Qi, J.X, Gao, G.F. | | 登録日 | 2023-12-28 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-08-07 | | 実験手法 | ELECTRON MICROSCOPY (3.31 Å) | | 主引用文献 | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 2024
|
|
6X0M
 
 | |
4KO3
 
 | | Low X-ray dose structure of anaerobically purified Dm. baculatum [NiFeSe]-hydrogenase after crystallization under air | | 分子名称: | CALCIUM ION, CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, ... | | 著者 | Volbeda, A, Cavazza, C, Fontecilla-Camps, J.C. | | 登録日 | 2013-05-11 | | 公開日 | 2013-07-10 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural foundations for the O2 resistance of Desulfomicrobium baculatum [NiFeSe]-hydrogenase.
Chem.Commun.(Camb.), 49, 2013
|
|
4KPJ
 
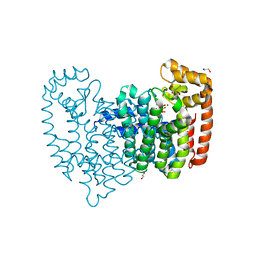 | | Crystal Structure of Farnesyl Pyrophosphate Synthase (Y204A) Mutant complexed with Mg, Pamidronate | | 分子名称: | 1,2-ETHANEDIOL, Farnesyl pyrophosphate synthase, MAGNESIUM ION, ... | | 著者 | Barnett, B.L, Tsoumpra, M.K, Muniz, J.R.C, Walter, R.L. | | 登録日 | 2013-05-13 | | 公開日 | 2014-04-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal Structure of Farnesyl Pyrophosphate Synthase (Y204A) Mutant complexed with Mg, Pamidronate
To be Published
|
|
4KQQ
 
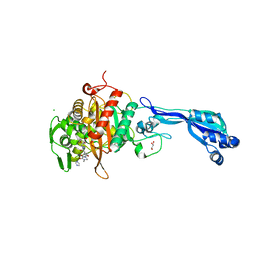 | | CRYSTAL STRUCTURE OF PENICILLIN-BINDING PROTEIN 3 FROM PSEUDOMONAS AERUGINOSA IN COMPLEX WITH (5S)-Penicilloic Acid | | 分子名称: | (2S,4S)-2-[(R)-carboxy{[(2R)-2-{[(4-ethyl-2,3-dioxopiperazin-1-yl)carbonyl]amino}-2-phenylacetyl]amino}methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | 著者 | Nettleship, J.E, Stuart, D.I, Owens, R.J, Ren, J. | | 登録日 | 2013-05-15 | | 公開日 | 2013-11-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Binding of (5S)-Penicilloic Acid to Penicillin Binding Protein 3.
Acs Chem.Biol., 8, 2013
|
|
4KML
 
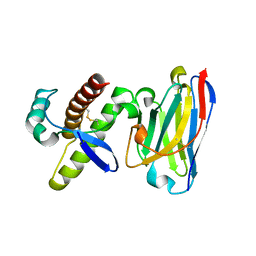 | | Probing the N-terminal beta-sheet conversion in the crystal structure of the full-length human prion protein bound to a Nanobody | | 分子名称: | Major prion protein, Nanobody | | 著者 | Abskharon, R.N.N, Giachin, G, Wohlkonig, A, Soror, S.H, Pardon, E, Legname, G, Steyaert, J. | | 登録日 | 2013-05-08 | | 公開日 | 2014-02-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Probing the N-Terminal beta-Sheet Conversion in the Crystal Structure of the Human Prion Protein Bound to a Nanobody.
J.Am.Chem.Soc., 136, 2014
|
|
4JZ6
 
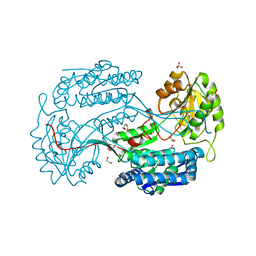 | | Crystal structure of a salicylaldehyde dehydrogenase from Pseudomonas putida G7 complexed with salicylaldehyde | | 分子名称: | 1,2-ETHANEDIOL, SALICYLALDEHYDE, SULFATE ION, ... | | 著者 | Coitinho, J.B, Nagem, R.A.P. | | 登録日 | 2013-04-02 | | 公開日 | 2014-04-02 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.417 Å) | | 主引用文献 | Structural and Kinetic Properties of the Aldehyde Dehydrogenase NahF, a Broad Substrate Specificity Enzyme for Aldehyde Oxidation.
Biochemistry, 55, 2016
|
|
4K11
 
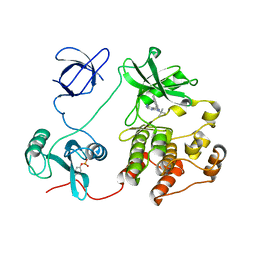 | | The structure of 1NA in complex with Src T338G | | 分子名称: | 1-tert-butyl-3-(naphthalen-1-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Proto-oncogene tyrosine-protein kinase Src | | 著者 | Eck, M.J, Yun, C.H. | | 登録日 | 2013-04-04 | | 公開日 | 2014-04-09 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The structure of 1NA in complex with Src T338G
To be Published
|
|
7C95
 
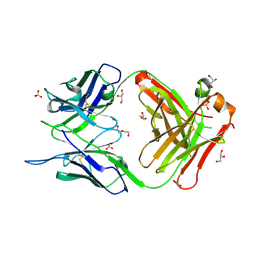 | | Crystal structure of the anti-human podoplanin antibody Fab fragment | | 分子名称: | GLYCEROL, Heavy chain of Fab fragment, Light chain of Fab fragment, ... | | 著者 | Nakamura, S, Suzuki, K, Ogasawara, S, Naruchi, K, Shimabukuro, J, Tukahara, N, Kaneko, M.K, Kato, Y, Murata, T. | | 登録日 | 2020-06-04 | | 公開日 | 2020-09-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Crystal structure of an anti-podoplanin antibody bound to a disialylated O-linked glycopeptide.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
4V3P
 
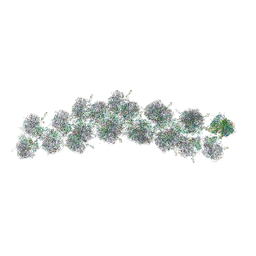 | | The molecular structure of the left-handed supra-molecular helix of eukaryotic polyribosomes | | 分子名称: | 18S ribosomal RNA, 26S ribosomal RNA, 40S WHEAT GERM RIBOSOME protein 4, ... | | 著者 | Myasnikov, A.G, Afonina, Z.A, Menetret, J.F, Shirokov, V.A, Spirin, A.S, Klaholz, B.P. | | 登録日 | 2014-10-20 | | 公開日 | 2015-04-22 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (34 Å) | | 主引用文献 | The molecular structure of the left-handed supra-molecular helix of eukaryotic polyribosomes.
Nat Commun, 5, 2014
|
|
4KQ8
 
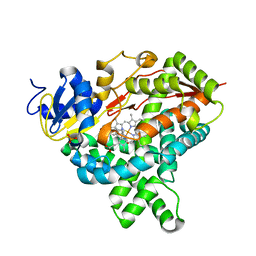 | | Structure of Recombinant Human Cytochrome P450 Aromatase | | 分子名称: | 4-ANDROSTENE-3-17-DIONE, Cytochrome P450 19A1, PHOSPHATE ION, ... | | 著者 | Ghosh, D, Di Nardo, G, Griswold, J. | | 登録日 | 2013-05-14 | | 公開日 | 2013-08-21 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.29 Å) | | 主引用文献 | Structural basis for the functional roles of critical residues in human cytochrome p450 aromatase.
Biochemistry, 52, 2013
|
|
