4YPB
 
 | | Precleavage 70S structure of the P. vulgaris HigB DeltaH92 toxin bound to the AAA codon | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Schureck, M.A, Dunkle, J.A, Maehigashi, T, Dunham, C.M. | | 登録日 | 2015-03-12 | | 公開日 | 2015-10-21 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Defining the mRNA recognition signature of a bacterial toxin protein.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6H58
 
 | | Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1 - Full 100S Hibernating E. coli Ribosome | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S1, ... | | 著者 | Beckert, B, Turk, M, Czech, A, Berninghausen, O, Beckmann, R, Ignatova, Z, Plitzko, J, Wilson, D.N. | | 登録日 | 2018-07-24 | | 公開日 | 2018-09-05 | | 最終更新日 | 2018-10-24 | | 実験手法 | ELECTRON MICROSCOPY (7.9 Å) | | 主引用文献 | Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1.
Nat Microbiol, 3, 2018
|
|
6GYS
 
 | | Cryo-EM structure of the CBF3-CEN3 complex of the budding yeast kinetochore | | 分子名称: | Centromere DNA-binding protein complex CBF3 subunit A, Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, ... | | 著者 | Yan, K, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | 登録日 | 2018-07-01 | | 公開日 | 2018-12-05 | | 最終更新日 | 2019-12-11 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Architecture of the CBF3-centromere complex of the budding yeast kinetochore.
Nat. Struct. Mol. Biol., 25, 2018
|
|
3JAI
 
 | | Structure of a mammalian ribosomal termination complex with ABCE1, eRF1(AAQ), and the UGA stop codon | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | 著者 | Brown, A, Shao, S, Murray, J, Hegde, R.S, Ramakrishnan, V. | | 登録日 | 2015-06-10 | | 公開日 | 2015-08-12 | | 最終更新日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (3.65 Å) | | 主引用文献 | Structural basis for stop codon recognition in eukaryotes.
Nature, 524, 2015
|
|
6HIW
 
 | | Cryo-EM structure of the Trypanosoma brucei mitochondrial ribosome - This entry contains the complete small mitoribosomal subunit in complex with mt-IF-3 | | 分子名称: | 9S rRNA, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Ramrath, D, Niemann, M, Leibundgut, M, Bieri, P, Prange, C, Horn, E.K, Leitner, A, Boehringer, D, Schneider, A, Ban, N. | | 登録日 | 2018-08-31 | | 公開日 | 2018-09-26 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.37 Å) | | 主引用文献 | Evolutionary shift toward protein-based architecture in trypanosomal mitochondrial ribosomes.
Science, 362, 2018
|
|
6R7Q
 
 | | Structure of XBP1u-paused ribosome nascent chain complex with Sec61. | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | 著者 | Shanmuganathan, V, Cheng, J, Braunger, K, Berninghausen, O, Beatrix, B, Beckmann, R. | | 登録日 | 2019-03-29 | | 公開日 | 2019-07-10 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural and mutational analysis of the ribosome-arresting human XBP1u.
Elife, 8, 2019
|
|
6RBD
 
 | | State 1 of yeast Tsr1-TAP Rps20-Deltaloop pre-40S particles | | 分子名称: | 20S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | 著者 | Shayan, R, Mitterer, V, Ferreira-Cerca, S, Murat, G, Enne, T, Rinaldi, D, Weigl, S, Omanic, H, Gleizes, P.E, Kressler, D, Pertschy, B, Plisson-Chastang, C. | | 登録日 | 2019-04-10 | | 公開日 | 2019-06-26 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.47 Å) | | 主引用文献 | Conformational proofreading of distant 40S ribosomal subunit maturation events by a long-range communication mechanism.
Nat Commun, 10, 2019
|
|
3K55
 
 | | Structure of beta hairpin deletion mutant of beta toxin from Staphylococcus aureus | | 分子名称: | Beta-hemolysin, CHLORIDE ION, SODIUM ION | | 著者 | Kruse, A.C, Huseby, M, Shi, K, Digre, J, Ohlendorf, D.H, Earhart, C.A. | | 登録日 | 2009-10-06 | | 公開日 | 2011-01-26 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.35 Å) | | 主引用文献 | Structure of a mutant beta toxin from Staphylococcus aureus reveals domain swapping and conformational flexibility
Acta Crystallogr.,Sect.F, 67, 2011
|
|
4Y20
 
 | |
4XBL
 
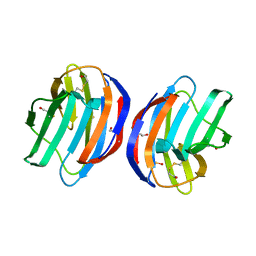 | |
8BQX
 
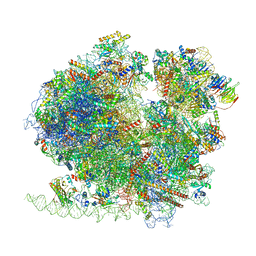 | | Yeast 80S ribosome in complex with Map1 (conformation 2) | | 分子名称: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | 著者 | Knorr, A.G, Mackens-Kiani, T, Musial, J, Berninghausen, O, Becker, T, Beatrix, B, Beckmann, R. | | 登録日 | 2022-11-21 | | 公開日 | 2023-03-22 | | 最終更新日 | 2023-05-03 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | The dynamic architecture of Map1- and NatB-ribosome complexes coordinates the sequential modifications of nascent polypeptide chains.
Plos Biol., 21, 2023
|
|
8BR8
 
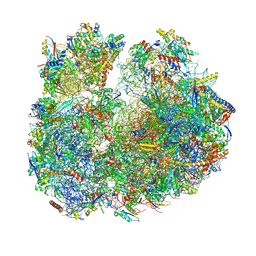 | | Giardia ribosome in POST-T state (A1) | | 分子名称: | 40S ribosomal protein S21, 40S ribosomal protein S25, 40S ribosomal protein S26, ... | | 著者 | Majumdar, S, Emmerich, A.G, Sanyal, S. | | 登録日 | 2022-11-22 | | 公開日 | 2023-03-15 | | 最終更新日 | 2023-05-03 | | 実験手法 | ELECTRON MICROSCOPY (3.35 Å) | | 主引用文献 | Insights into translocation mechanism and ribosome evolution from cryo-EM structures of translocation intermediates of Giardia intestinalis.
Nucleic Acids Res., 51, 2023
|
|
4XID
 
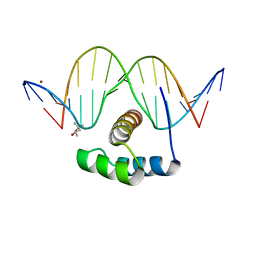 | | AntpHD with 15bp DNA duplex | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*AP*GP*AP*AP*AP*GP*CP*CP*AP*TP*TP*AP*GP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*CP*TP*AP*AP*TP*GP*GP*CP*TP*TP*TP*C)-3'), ... | | 著者 | White, M.A, Zandarashvili, L, Iwahara, J. | | 登録日 | 2015-01-06 | | 公開日 | 2015-11-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.701 Å) | | 主引用文献 | Entropic Enhancement of Protein-DNA Affinity by Oxygen-to-Sulfur Substitution in DNA Phosphate.
Biophys.J., 109, 2015
|
|
8BUU
 
 | | ARE-ABCF VmlR2 bound to a 70S ribosome | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Crowe-McAuliffe, C, Wilson, D.N. | | 登録日 | 2022-11-30 | | 公開日 | 2023-04-05 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Genome-encoded ABCF factors implicated in intrinsic antibiotic resistance in Gram-positive bacteria: VmlR2, Ard1 and CplR.
Nucleic Acids Res., 51, 2023
|
|
6Q6G
 
 | | Cryo-EM structure of the APC/C-Cdc20-Cdk2-cyclinA2-Cks2 complex, the D1 box class | | 分子名称: | Anaphase-promoting complex subunit 1,Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | 著者 | Zhang, S, Barford, D. | | 登録日 | 2018-12-11 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cyclin A2 degradation during the spindle assembly checkpoint requires multiple binding modes to the APC/C.
Nat Commun, 10, 2019
|
|
6Q98
 
 | |
7YE1
 
 | | The cryo-EM structure of C. crescentus GcrA-TACup | | 分子名称: | Cell cycle regulatory protein GcrA, DNA (57-MER)-non template, DNA (57-MER)-template, ... | | 著者 | Wu, X.X, Zhang, Y. | | 登録日 | 2022-07-05 | | 公開日 | 2023-01-18 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cryo-EM structures of Caulobacter crescentus transcription activation complex with an essential cell cycle regulator GcrA
Nucleic Acids Res., 2023
|
|
7YLA
 
 | | Cryo-EM structure of 50S-HflX complex | | 分子名称: | 50S ribosomal protein L11, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | 著者 | Damu, W, Ning, G. | | 登録日 | 2022-07-25 | | 公開日 | 2023-01-04 | | 最終更新日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (2.52 Å) | | 主引用文献 | Cryo-EM Structure of the 50S-HflX Complex Reveals a Novel Mechanism of Antibiotic Resistance in E. coli
Biorxiv, 2022
|
|
6G53
 
 | | Cryo-EM structure of a late human pre-40S ribosomal subunit - State E | | 分子名称: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | 著者 | Ameismeier, M, Cheng, J, Berninghausen, O, Beckmann, R. | | 登録日 | 2018-03-28 | | 公開日 | 2018-06-06 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Visualizing late states of human 40S ribosomal subunit maturation.
Nature, 558, 2018
|
|
7Y4L
 
 | | PBS of PBS-PSII-PSI-LHCs from Porphyridium purpureum. | | 分子名称: | Allophycocyanin alpha subunit, Allophycocyanin beta 18 subunit, Allophycocyanin beta subunit, ... | | 著者 | You, X, Zhang, X, Cheng, J, Xiao, Y.N, Sun, S, Sui, S.F. | | 登録日 | 2022-06-15 | | 公開日 | 2023-01-18 | | 最終更新日 | 2023-04-19 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | In situ structure of the red algal phycobilisome-PSII-PSI-LHC megacomplex.
Nature, 616, 2023
|
|
8A5I
 
 | | Cryo-EM structure of Lincomycin bound to the Listeria monocytogenes 50S ribosomal subunit. | | 分子名称: | 1,4-DIAMINOBUTANE, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | 著者 | Koller, T.O, Crowe-McAuliffe, C, Wilson, D.N. | | 登録日 | 2022-06-15 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.3 Å) | | 主引用文献 | Structural basis for HflXr-mediated antibiotic resistance in Listeria monocytogenes.
Nucleic Acids Res., 50, 2022
|
|
8A63
 
 | | Cryo-EM structure of Listeria monocytogenes 50S ribosomal subunit. | | 分子名称: | 1,4-DIAMINOBUTANE, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | 著者 | Koller, T.O, Crowe-McAuliffe, C, Wilson, D.N. | | 登録日 | 2022-06-16 | | 公開日 | 2022-11-02 | | 最終更新日 | 2022-11-16 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis for HflXr-mediated antibiotic resistance in Listeria monocytogenes.
Nucleic Acids Res., 50, 2022
|
|
6FVV
 
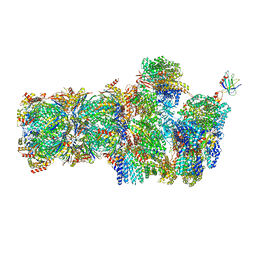 | | 26S proteasome, s3 state | | 分子名称: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, ... | | 著者 | Eisele, M.R, Reed, R.G, Rudack, T, Schweitzer, A, Beck, F, Nagy, I, Pfeifer, G, Plitzko, J.M, Baumeister, W, Tomko, R.J, Sakata, E. | | 登録日 | 2018-03-05 | | 公開日 | 2018-08-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (5.4 Å) | | 主引用文献 | Expanded Coverage of the 26S Proteasome Conformational Landscape Reveals Mechanisms of Peptidase Gating.
Cell Rep, 24, 2018
|
|
8A57
 
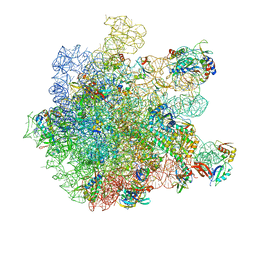 | | Cryo-EM structure of HflXr bound to the Listeria monocytogenes 50S ribosomal subunit. | | 分子名称: | 1,4-DIAMINOBUTANE, 23S ribosomal RNA, 50S ribosomal protein L11, ... | | 著者 | Koller, T.O, Crowe-McAuliffe, C, Wilson, D.N. | | 登録日 | 2022-06-14 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.3 Å) | | 主引用文献 | Structural basis for HflXr-mediated antibiotic resistance in Listeria monocytogenes.
Nucleic Acids Res., 50, 2022
|
|
6PWE
 
 | |
