4ITA
 
 | | Structure of bacterial enzyme in complex with cofactor | | 分子名称: | 1,2-ETHANEDIOL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Succinate-semialdehyde dehydrogenase | | 著者 | Rhee, S, Park, J. | | 登録日 | 2013-01-18 | | 公開日 | 2013-04-24 | | 最終更新日 | 2013-06-26 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural Basis for a Cofactor-dependent Oxidation Protection and Catalysis of Cyanobacterial Succinic Semialdehyde Dehydrogenase.
J.Biol.Chem., 288, 2013
|
|
3O88
 
 | | Crystal structure of AmpC beta-lactamase in complex with a sulfonamide boronic acid inhibitor | | 分子名称: | 3-[(2R)-2-[(benzylsulfonyl)amino]-2-(dihydroxyboranyl)ethyl]benzoic acid, Beta-lactamase, PHOSPHATE ION | | 著者 | Eidam, O, Romagnoli, C, Karpiak, J, Shoichet, B.K. | | 登録日 | 2010-08-02 | | 公開日 | 2010-11-03 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Design, Synthesis, Crystal Structures, and Antimicrobial Activity of Sulfonamide Boronic Acids as beta-Lactamase Inhibitors
J.Med.Chem., 53, 2010
|
|
3O8S
 
 | |
2HYK
 
 | | The crystal structure of an endo-beta-1,3-glucanase from alkaliphilic Nocardiopsis sp.strain F96 | | 分子名称: | Beta-1,3-glucanase, CALCIUM ION, ETHANOL, ... | | 著者 | Fibriansah, G, Nakamura, S, Kumasaka, T. | | 登録日 | 2006-08-07 | | 公開日 | 2007-10-02 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | The 1.3 A crystal structure of a novel endo-beta-1,3-glucanase of glycoside hydrolase family 16 from alkaliphilic Nocardiopsis sp. strain F96.
Proteins, 69, 2007
|
|
4IT3
 
 | |
3O9D
 
 | | Crystal Structure of wild-type HIV-1 Protease in complex with kd19 | | 分子名称: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-3-{(1,3-benzothiazol-6-ylsulfonyl)[(2S)-2-methylbutyl]amino}-1-benzyl-2-hydroxypropyl]carbamate, ACETATE ION, PHOSPHATE ION, ... | | 著者 | Schiffer, C.A, Nalam, M.N.L. | | 登録日 | 2010-08-04 | | 公開日 | 2011-08-10 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Substrate envelope-designed potent HIV-1 protease inhibitors to avoid drug resistance.
Chem.Biol., 20, 2013
|
|
2HYZ
 
 | |
4IT9
 
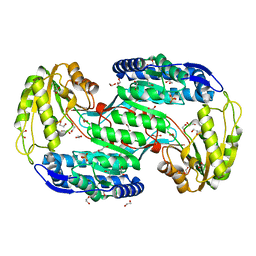 | | Structure of Bacterial Enzyme | | 分子名称: | 1,2-ETHANEDIOL, PHOSPHATE ION, Succinate-semialdehyde dehydrogenase | | 著者 | Rhee, S, Park, J. | | 登録日 | 2013-01-18 | | 公開日 | 2013-04-24 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural Basis for a Cofactor-dependent Oxidation Protection and Catalysis of Cyanobacterial Succinic Semialdehyde Dehydrogenase.
J.Biol.Chem., 288, 2013
|
|
2P84
 
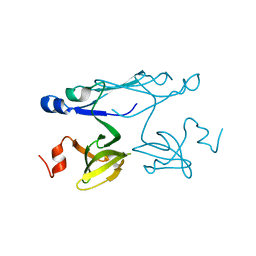 | | Crystal structure of ORF041 from Bacteriophage 37 | | 分子名称: | ORF041 | | 著者 | Malashkevich, V.N, Toro, R, Sauder, J.M, Schwinn, K.D, Thompson, D.A, Bain, K.T, Adams, J.M, Reyes, C, Lau, C, Gilmore, J, Rooney, I, Wasserman, T, Gheyi, S.R, Emtage, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2007-03-21 | | 公開日 | 2007-04-03 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of the hypothetical protein from Staphylococcus phage 37
To be Published
|
|
3GPI
 
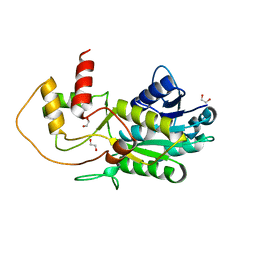 | | Structure of putative NAD-dependent epimerase/dehydratase from methylobacillus flagellatus | | 分子名称: | 1,2-ETHANEDIOL, NAD-dependent epimerase/dehydratase | | 著者 | Ramagopal, U.A, Morano, C, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-03-23 | | 公開日 | 2009-04-21 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.44 Å) | | 主引用文献 | Structure of putative NAD-dependent epimerase/dehydratase from methylobacillus flagellatus
To be published
|
|
2PEN
 
 | | Crystal structure of RbcX, crystal form I | | 分子名称: | ORF134 | | 著者 | Saschenbrecker, S, Bracher, A, Vasudeva Rao, K, Vasudeva Rao, B, Hartl, F.U, Hayer-Hartl, M. | | 登録日 | 2007-04-03 | | 公開日 | 2007-07-10 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure and Function of RbcX, an Assembly Chaperone for Hexadecameric Rubisco.
Cell(Cambridge,Mass.), 129, 2007
|
|
3O9I
 
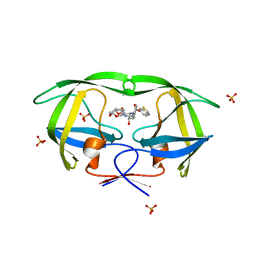 | | Crystal Structure of wild-type HIV-1 Protease in complex with af61 | | 分子名称: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(1S,2R)-3-[(1,3-benzothiazol-6-ylsulfonyl)(2-ethylbutyl)amino]-1-benzyl-2-hydroxypropyl}carbamate, PHOSPHATE ION, Protease | | 著者 | Schiffer, C.A, Nalam, M.N.L. | | 登録日 | 2010-08-04 | | 公開日 | 2011-08-10 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Substrate envelope-designed potent HIV-1 protease inhibitors to avoid drug resistance.
Chem.Biol., 20, 2013
|
|
3GR4
 
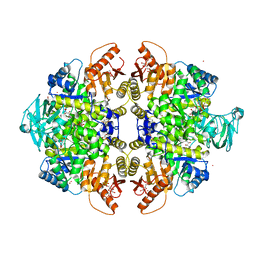 | | Activator-Bound Structure of Human Pyruvate Kinase M2 | | 分子名称: | 1,6-di-O-phosphono-beta-D-fructofuranose, 1-[(2,6-difluorophenyl)sulfonyl]-4-(2,3-dihydro-1,4-benzodioxin-6-ylsulfonyl)piperazine, ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Hong, B, Dimov, S, Tempel, W, Auld, D, Thomas, C, Boxer, M, Jianq, J.-K, Skoumbourdis, A, Min, S, Southall, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Inglese, J, Park, H, Structural Genomics Consortium (SGC) | | 登録日 | 2009-03-24 | | 公開日 | 2009-04-07 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Activator-Bound Structures of Human Pyruvate Kinase M2
to be published
|
|
3ODA
 
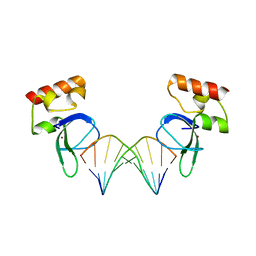 | | Human PARP-1 zinc finger 1 (Zn1) bound to DNA | | 分子名称: | 5'-D(*GP*CP*CP*TP*GP*CP*AP*GP*GP*C)-3', Poly [ADP-ribose] polymerase 1, ZINC ION | | 著者 | Pascal, J.M, Langelier, M.-F. | | 登録日 | 2010-08-11 | | 公開日 | 2011-01-12 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.64 Å) | | 主引用文献 | Crystal Structures of Poly(ADP-ribose) Polymerase-1 (PARP-1) Zinc Fingers Bound to DNA: STRUCTURAL AND FUNCTIONAL INSIGHTS INTO DNA-DEPENDENT PARP-1 ACTIVITY.
J.Biol.Chem., 286, 2011
|
|
3OBT
 
 | |
2ONV
 
 | |
4ONL
 
 | | Crystal structure of human Mms2/Ubc13_D81N, R85S, A122V, N123P | | 分子名称: | Ubiquitin-conjugating enzyme E2 N, Ubiquitin-conjugating enzyme E2 variant 2 | | 著者 | Hodge, C.D, Edwards, R.A, Glover, J.N.M. | | 登録日 | 2014-01-28 | | 公開日 | 2015-05-06 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Covalent Inhibition of Ubc13 Affects Ubiquitin Signaling and Reveals Active Site Elements Important for Targeting.
Acs Chem.Biol., 10, 2015
|
|
4IWN
 
 | | Crystal structure of a putative methyltransferase CmoA in complex with a novel SAM derivative | | 分子名称: | (2S)-4-[{[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}(carboxylatomethyl)sulfonio] -2-ammoniobutanoate, (4S)-2-METHYL-2,4-PENTANEDIOL, tRNA (cmo5U34)-methyltransferase | | 著者 | Aller, P, Lobley, C.M, Byrne, R.T, Antson, A.A, Waterman, D.G. | | 登録日 | 2013-01-24 | | 公開日 | 2013-05-29 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | S-Adenosyl-S-carboxymethyl-L-homocysteine: a novel cofactor found in the putative tRNA-modifying enzyme CmoA.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3OCY
 
 | | Structure of Recombinant Haemophilus Influenzae e(P4) Acid Phosphatase Complexed with inorganic phosphate | | 分子名称: | Lipoprotein E, MAGNESIUM ION, PHOSPHATE ION | | 著者 | Singh, H, Schuermann, J, Reilly, T, Calcutt, M, Tanner, J. | | 登録日 | 2010-08-10 | | 公開日 | 2010-10-20 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Recognition of nucleoside monophosphate substrates by Haemophilus influenzae class C acid phosphatase.
J.Mol.Biol., 404, 2010
|
|
3OEO
 
 | | The crystal structure E. coli Spy | | 分子名称: | CADMIUM ION, Spheroplast protein Y | | 著者 | Kwon, E, Kim, D.Y, Gross, C.A, Gross, J.D, Kim, K.K. | | 登録日 | 2010-08-13 | | 公開日 | 2010-09-22 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | The crystal structure Escherichia coli Spy.
Protein Sci., 19, 2010
|
|
3OD9
 
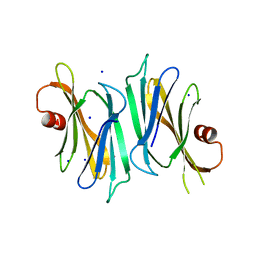 | | Crystal structure of PliI-Ah, periplasmic lysozyme inhibitor of I-type lysozyme from Aeromonas hydrophyla | | 分子名称: | POTASSIUM ION, Putative exported protein, SODIUM ION | | 著者 | Leysen, S, Van Herreweghe, J.M, Callewaert, L, Heirbaut, M, Buntinx, P, Michiels, C.W, Strelkov, S.V. | | 登録日 | 2010-08-11 | | 公開日 | 2010-12-22 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.411 Å) | | 主引用文献 | Molecular Basis of Bacterial Defense against Host Lysozymes: X-ray Structures of Periplasmic Lysozyme Inhibitors PliI and PliC.
J.Mol.Biol., 405, 2011
|
|
2PBY
 
 | | Probable Glutaminase from Geobacillus kaustophilus HTA426 | | 分子名称: | Glutaminase | | 著者 | Dillard, B.D, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Rose, J.P, Wang, B.-C, RIKEN Structural Genomics/Proteomics Initiative (RSGI), Southeast Collaboratory for Structural Genomics (SECSG) | | 登録日 | 2007-03-29 | | 公開日 | 2007-06-12 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Glutaminase from Geobacillus kaustophilus HTA426
To be Published
|
|
3GVV
 
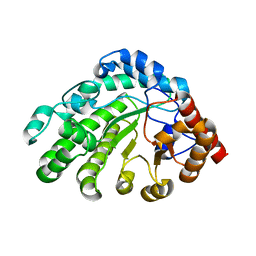 | | Single-chain UROD Y164G (GY) mutation | | 分子名称: | Uroporphyrinogen decarboxylase | | 著者 | Hill, C.P, Phillips, J.D, Whitby, F.G, Warby, C, Kushner, J.P. | | 登録日 | 2009-03-31 | | 公開日 | 2009-07-07 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Substrate shuttling between active sites of uroporphyrinogen decarboxylase is not required to generate coproporphyrinogen.
J.Mol.Biol., 389, 2009
|
|
3OGD
 
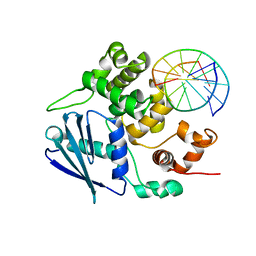 | | AlkA Undamaged DNA Complex: Interrogation of a G*:C base pair | | 分子名称: | 5'-D(*CP*AP*(BRU)P*GP*AP*CP*(BRU)P*GP*C)-3', 5'-D(*GP*CP*AP*GP*TP*CP*AP*TP*G)-3', DNA-3-methyladenine glycosylase 2 | | 著者 | Bowman, B.R, Lee, S, Wang, S, Verdine, G.L. | | 登録日 | 2010-08-16 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of Escherichia coli AlkA in Complex with Undamaged DNA.
J.Biol.Chem., 285, 2010
|
|
3GPP
 
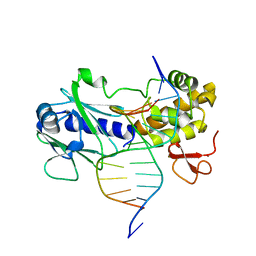 | | MutM encountering an intrahelical 8-oxoguanine (oxoG) lesion in EC3-T224P complex | | 分子名称: | DNA (5'-D(P*CP*GP*TP*CP*CP*(8OG)P*GP*AP*TP*CP*TP*AP*C)-3'), DNA (5'-D(P*GP*GP*TP*AP*GP*AP*TP*CP*CP*GP*GP*AP*C)-3'), DNA glycosylase, ... | | 著者 | Spong, M.C, Qi, Y, Verdine, G.L. | | 登録日 | 2009-03-23 | | 公開日 | 2009-11-10 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Encounter and extrusion of an intrahelical lesion by a DNA repair enzyme.
Nature, 462, 2009
|
|
