4Z6E
 
 | |
7AN7
 
 | | Enzyme of biosynthetic pathway | | 分子名称: | 3-[(1-Carboxyvinyl)oxy]benzoic acid, Chorismate dehydratase, GLYCEROL | | 著者 | Archna, A, Breithaupt, C, Stubbs, M.T. | | 登録日 | 2020-10-11 | | 公開日 | 2022-04-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Mechanism of chorismate dehydratase MqnA, the first enzyme of the futalosine pathway, proceeds via substrate-assisted catalysis
J.Biol.Chem., 2022
|
|
7AN8
 
 | | Enzyme of biosynthetic pathway | | 分子名称: | 3-[(1-Carboxyvinyl)oxy]benzoic acid, Chorismate dehydratase, GLYCEROL | | 著者 | Archna, A, Breithaupt, C, Stubbs, M.T. | | 登録日 | 2020-10-11 | | 公開日 | 2022-04-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Mechanism of chorismate dehydratase MqnA, the first enzyme of the futalosine pathway, proceeds via substrate-assisted catalysis
J.Biol.Chem., 2022
|
|
7AN9
 
 | |
7B1T
 
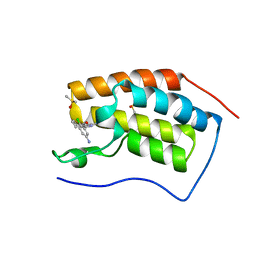 | | Crystal structure of BRD4(1) in complex with the inhibitor MPM6 | | 分子名称: | 3-(5-azanyl-2-chloranyl-phenyl)-1-methyl-4,7-dihydro-2~{H}-cyclohepta[c]pyrrol-8-one, Bromodomain-containing protein 4, DIMETHYL SULFOXIDE | | 著者 | Huegle, M. | | 登録日 | 2020-11-25 | | 公開日 | 2022-06-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | A novel pan-selective bromodomain inhibitor for epigenetic drug design.
Eur.J.Med.Chem., 249, 2023
|
|
4YAW
 
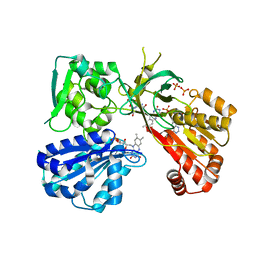 | | Reduced CYPOR mutant - G141del | | 分子名称: | ADENOSINE-2'-MONOPHOSPHATE, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Xia, C, Kim, J.J.P. | | 登録日 | 2015-02-17 | | 公開日 | 2016-03-02 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Mutants of Cytochrome P450 Reductase Lacking Either Gly-141 or Gly-143 Destabilize Its FMN Semiquinone.
J.Biol.Chem., 291, 2016
|
|
5E6E
 
 | |
6BOS
 
 | |
5FF6
 
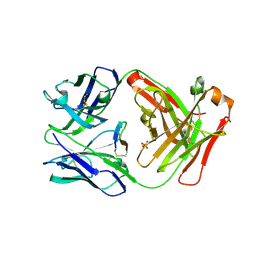 | | Cetuximab Fab in complex with L10Q meditope variant | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cetuximab Fab heavy chain, Cetuximab Fab light chain, ... | | 著者 | Bzymek, K.P, Williams, J.C. | | 登録日 | 2015-12-17 | | 公開日 | 2016-10-26 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Natural and non-natural amino-acid side-chain substitutions: affinity and diffraction studies of meditope-Fab complexes.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
6BVE
 
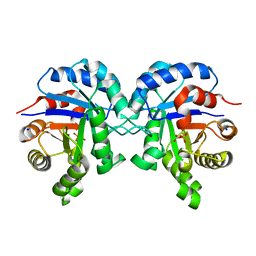 | |
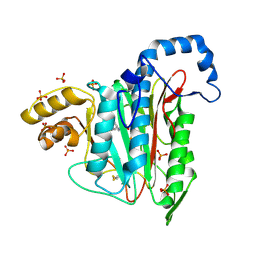
7A12
 
 | |
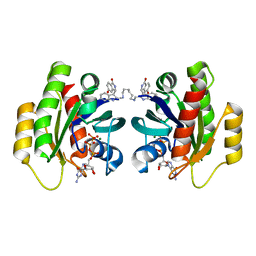
7ACH
 
 | |
4YAU
 
 | | Reduced CYPOR mutant - G141del/E142N | | 分子名称: | ADENOSINE-2'-MONOPHOSPHATE, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Xia, C, Kim, J.J.P. | | 登録日 | 2015-02-17 | | 公開日 | 2016-03-02 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Mutants of Cytochrome P450 Reductase Lacking Either Gly-141 or Gly-143 Destabilize Its FMN Semiquinone.
J.Biol.Chem., 291, 2016
|
|
6BMB
 
 | | Crystal structure of Arabidopsis Dehydroquinate dehydratase-shikimate dehydrogenase (T381G mutant) in complex with tartrate and shikimate | | 分子名称: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, Bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase, chloroplastic, ... | | 著者 | Christendat, D, Peek, J. | | 登録日 | 2017-11-14 | | 公開日 | 2017-12-20 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.077 Å) | | 主引用文献 | Structural and biochemical approaches uncover multiple evolutionary trajectories of plant quinate dehydrogenases.
Plant J., 2018
|
|
4YFJ
 
 | | Crystal structure of aminoglycoside acetyltransferase AAC(3)-Ib | | 分子名称: | Aminoglycoside 3'-N-acetyltransferase, SULFATE ION | | 著者 | Stogios, P.J, Xu, Z, Evdokimova, E, Yim, V, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-02-25 | | 公開日 | 2015-03-18 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of aminoglycoside acetyltransferase AAC(3)-Ib
To Be Published
|
|
6BIQ
 
 | |
6BMQ
 
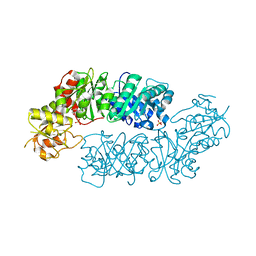 | | Crystal structure of Arabidopsis Dehydroquinate dehydratase-shikimate dehydrogenase (T381G mutant) in complex with tartrate and shikimate | | 分子名称: | (1S,3R,4S,5R)-1,3,4,5-tetrahydroxycyclohexanecarboxylic acid, Bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase, chloroplastic, ... | | 著者 | Christendat, D, Peek, J. | | 登録日 | 2017-11-15 | | 公開日 | 2018-09-26 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2.077 Å) | | 主引用文献 | Structural and biochemical approaches uncover multiple evolutionary trajectories of plant quinate dehydrogenases.
Plant J., 2018
|
|
7BHO
 
 | | DNA origami signpost designed model | | 分子名称: | DNA, DNA scaffold | | 著者 | Silvester, E, Vollmer, B, Prazak, V, Vasishtan, D, Machala, E.A, Whittle, C, Black, S, Bath, J, Turberfield, A.J, Gruenewald, K, Baker, L.A. | | 登録日 | 2021-01-11 | | 公開日 | 2021-04-14 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (36.439999 Å) | | 主引用文献 | DNA origami signposts for identifying proteins on cell membranes by electron cryotomography.
Cell, 184, 2021
|
|
5F49
 
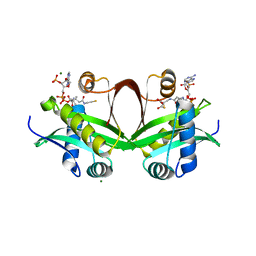 | | Crystal structure of an aminoglycoside acetyltransferase meta-AAC0020 from an uncultured soil metagenomic sample in complex with malonyl-coenzyme A | | 分子名称: | COENZYME A, MAGNESIUM ION, MALONYL-COENZYME A, ... | | 著者 | Xu, Z, Skarina, T, Stogios, P.J, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-12-03 | | 公開日 | 2015-12-16 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structural and Functional Survey of Environmental Aminoglycoside Acetyltransferases Reveals Functionality of Resistance Enzymes.
ACS Infect Dis, 3, 2017
|
|
6BJR
 
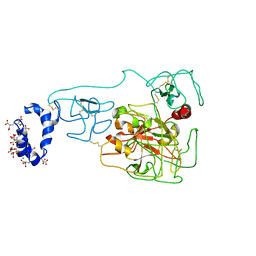 | | Crystal structure of prothrombin mutant S101C/A470C | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Prothrombin, ... | | 著者 | Chinnaraj, M, Chen, Z, Pelc, L, Grese, Z, Bystranowska, D, Di Cera, E, Pozzi, N. | | 登録日 | 2017-11-06 | | 公開日 | 2018-06-27 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (6 Å) | | 主引用文献 | Structure of prothrombin in the closed form reveals new details on the mechanism of activation.
Sci Rep, 8, 2018
|
|
7BJ3
 
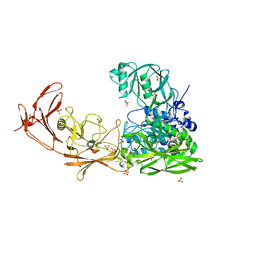 | | ScpA from Streptococcus pyogenes, S512A active site mutant | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, C5a peptidase, CALCIUM ION, ... | | 著者 | Kagawa, T.F, O'Connell, M.R, Cooney, J.C. | | 登録日 | 2021-01-13 | | 公開日 | 2021-05-12 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Enzyme kinetic and binding studies identify determinants of specificity for the immunomodulatory enzyme ScpA, a C5a inactivating bacterial protease.
Comput Struct Biotechnol J, 19, 2021
|
|
5ETU
 
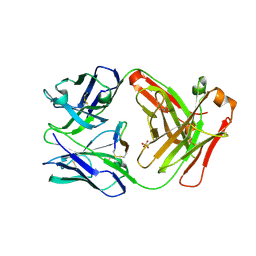 | | Cetuximab Fab in complex with L5E meditope variant | | 分子名称: | Cetuximab Fab heavy chain, Cetuximab Fab light chain, L5E meditope variant, ... | | 著者 | Bzymek, K.P, Williams, J.C. | | 登録日 | 2015-11-18 | | 公開日 | 2016-10-26 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.53 Å) | | 主引用文献 | Natural and non-natural amino-acid side-chain substitutions: affinity and diffraction studies of meditope-Fab complexes.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
4YAO
 
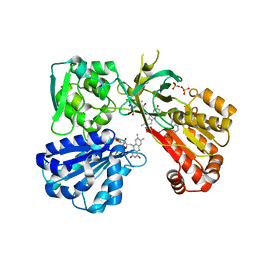 | | Reduced CYPOR mutant - G143del | | 分子名称: | ADENOSINE-2'-MONOPHOSPHATE, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Xia, C, Kim, J.J.P. | | 登録日 | 2015-02-17 | | 公開日 | 2016-03-02 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Mutants of Cytochrome P450 Reductase Lacking Either Gly-141 or Gly-143 Destabilize Its FMN Semiquinone.
J.Biol.Chem., 291, 2016
|
|
7BGQ
 
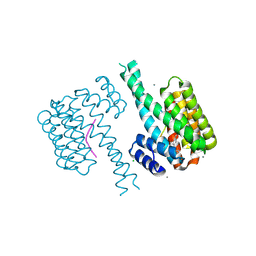 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound LvD1019 | | 分子名称: | 14-3-3 protein sigma, 2-methoxy-4-(2-phenylimidazol-1-yl)benzaldehyde, CALCIUM ION, ... | | 著者 | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | 登録日 | 2021-01-08 | | 公開日 | 2021-06-16 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
5F6W
 
 | | Crystal structure of Ubc9 (K48/K49A/E54A) complexed with Fragment 1 (biphenol) | | 分子名称: | 2-(2-hydroxyphenyl)phenol, SUMO-conjugating enzyme UBC9 | | 著者 | Lountos, G.T, Hewitt, W.M, Zlotkowski, K, Dahlhauser, S, Saunders, L.B, Needle, D, Tropea, J.E, Zhan, C, Wei, G, Ma, B, Nussinov, R, Schneekloth, J.S.Jr, Waugh, D.S. | | 登録日 | 2015-12-07 | | 公開日 | 2016-04-27 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.699 Å) | | 主引用文献 | Insights Into the Allosteric Inhibition of the SUMO E2 Enzyme Ubc9.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
