1LA3
 
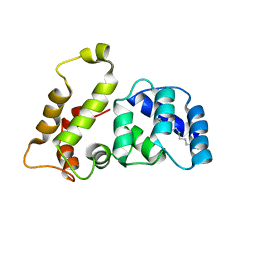 | | Solution structure of recoverin mutant, E85Q | | 分子名称: | CALCIUM ION, MYRISTIC ACID, Recoverin | | 著者 | Ames, J.B, Hamasaki, N, Molchanova, T. | | 登録日 | 2002-03-27 | | 公開日 | 2002-06-19 | | 最終更新日 | 2021-10-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure and calcium-binding studies of a recoverin mutant (E85Q) in an allosteric intermediate state.
Biochemistry, 41, 2002
|
|
6SEI
 
 | | Recognition and processing of branched DNA substrates by Slx1-Slx4 nuclease | | 分子名称: | CALCIUM ION, DNA (32-MER), Structure-specific endonuclease subunit SLX1, ... | | 著者 | Gaur, V, Zajko, W, Nirwal, S, Szlachcic, A, Gapinska, M, Nowotny, M. | | 登録日 | 2019-07-30 | | 公開日 | 2019-09-25 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Recognition and processing of branched DNA substrates by Slx1-Slx4 nuclease.
Nucleic Acids Res., 47, 2019
|
|
6UZD
 
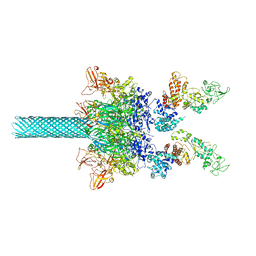 | | Anthrax toxin protective antigen channels bound to edema factor | | 分子名称: | CALCIUM ION, Calmodulin-sensitive adenylate cyclase, Protective antigen | | 著者 | Hardenbrook, N.J, Liu, S, Zhou, K, Zhou, Z.H, Krantz, B.A. | | 登録日 | 2019-11-14 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Atomic structures of anthrax toxin protective antigen channels bound to partially unfolded lethal and edema factors.
Nat Commun, 11, 2020
|
|
3KQH
 
 | |
5YQQ
 
 | |
3K52
 
 | |
5WK0
 
 | |
4ELD
 
 | |
3OPZ
 
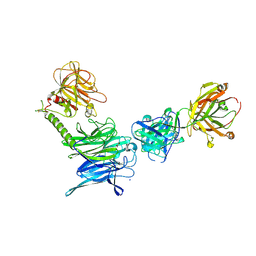 | | Crystal structure of trans-sialidase in complex with the Fab fragment of a neutralizing monoclonal IgG antibody | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, SODIUM ION, Trans-sialidase, ... | | 著者 | Larrieux, N, Muia, R, Campetella, O, Buschiazzo, A. | | 登録日 | 2010-09-02 | | 公開日 | 2011-11-09 | | 最終更新日 | 2021-10-06 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Trypanosoma cruzi trans-sialidase in complex with a neutralizing antibody: structure/function studies towards the rational design of inhibitors.
Plos Pathog., 8, 2012
|
|
3P3I
 
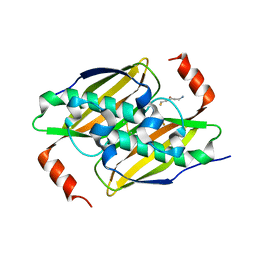 | | Crystal structure of the F36A mutant of the fluoroacetyl-CoA-specific thioesterase FlK in complex with fluoroacetate and CoA | | 分子名称: | COENZYME A, Fluoroacetyl coenzyme A thioesterase, fluoroacetic acid | | 著者 | Weeks, A.M, Coyle, S.M, Jinek, M, Doudna, J.A, Chang, M.C.Y. | | 登録日 | 2010-10-04 | | 公開日 | 2010-10-20 | | 最終更新日 | 2011-11-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural and biochemical studies of a fluoroacetyl-CoA-specific thioesterase reveal a molecular basis for fluorine selectivity.
Biochemistry, 49, 2010
|
|
2C9T
 
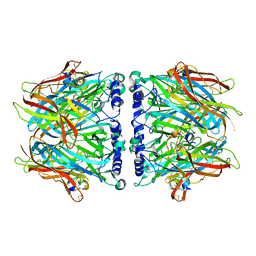 | | Crystal Structure Of Acetylcholine Binding Protein (AChBP) From Aplysia Californica In Complex With alpha-Conotoxin ImI | | 分子名称: | ALPHA-CONOTOXIN IMI, SOLUBLE ACETYLCHOLINE RECEPTOR | | 著者 | Ulens, C, Hogg, R.C, Celie, P.H, Bertrand, D, Tsetlin, V, Smit, A.B, Sixma, T.K. | | 登録日 | 2005-12-14 | | 公開日 | 2006-02-13 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structural Determinants of Selective {Alpha}-Conotoxin Binding to a Nicotinic Acetylcholine Receptor Homolog Achbp.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
5NG0
 
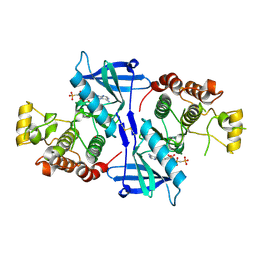 | | Structure of RIP2K(L294F) with bound AMPPCP | | 分子名称: | COBALT (II) ION, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | 著者 | Pellegrini, E, Cusack, S. | | 登録日 | 2017-03-16 | | 公開日 | 2017-06-07 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structures of the inactive and active states of RIP2 kinase inform on the mechanism of activation.
PLoS ONE, 12, 2017
|
|
5NG3
 
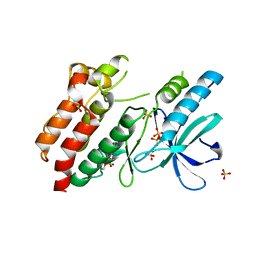 | | Structure of inactive kinase RIP2K(K47R) | | 分子名称: | Receptor-interacting serine/threonine-protein kinase 2, SULFATE ION | | 著者 | Pellegrini, E, Cusack, S. | | 登録日 | 2017-03-16 | | 公開日 | 2017-06-28 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structures of the inactive and active states of RIP2 kinase inform on the mechanism of activation.
PLoS ONE, 12, 2017
|
|
5YQI
 
 | | Crystal structure of the first StARkin domain of Lam2 | | 分子名称: | Membrane-anchored lipid-binding protein YSP2 | | 著者 | Tong, J, Im, Y.J. | | 登録日 | 2017-11-06 | | 公開日 | 2018-01-31 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural basis of sterol recognition and nonvesicular transport by lipid transfer proteins anchored at membrane contact sites
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5NOF
 
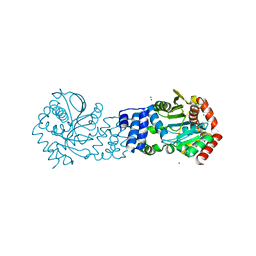 | | Anthranilate phosphoribosyltransferase from Thermococcus kodakaraensis | | 分子名称: | Anthranilate phosphoribosyltransferase, CHLORIDE ION, SODIUM ION, ... | | 著者 | Perveen, S, Rashid, N, Papageorgiou, A.C. | | 登録日 | 2017-04-12 | | 公開日 | 2017-11-29 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.42 Å) | | 主引用文献 | Anthranilate phosphoribosyltransferase from the hyperthermophilic archaeon Thermococcus kodakarensis shows maximum activity with zinc and forms a unique dimeric structure.
FEBS Open Bio, 7, 2017
|
|
1JDI
 
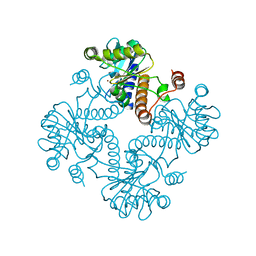 | | CRYSTAL STRUCTURE OF L-RIBULOSE-5-PHOSPHATE 4-EPIMERASE | | 分子名称: | L-RIBULOSE 5 PHOSPHATE 4-EPIMERASE, ZINC ION | | 著者 | Luo, Y, Samuel, J, Mosimann, S.C, Lee, J.E, Tanner, M.E, Strynadka, N.C.J. | | 登録日 | 2001-06-13 | | 公開日 | 2002-01-23 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The structure of L-ribulose-5-phosphate 4-epimerase: an aldolase-like platform for epimerization.
Biochemistry, 40, 2001
|
|
2XGD
 
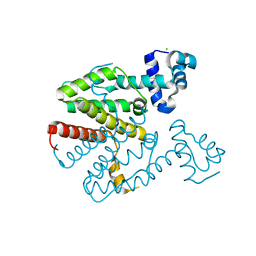 | | Crystal structure of a designed homodimeric variant T-A(L)A(L) of the tetracycline repressor | | 分子名称: | CHLORIDE ION, TETRACYCLINE REPRESSOR PROTEIN CLASS B FROM TRANSPOSON TN10, TETRACYCLINE REPRESSOR PROTEIN CLASS D | | 著者 | Stiebritz, M.T, Wengrzik, S, Richter, J.P, Muller, Y.A. | | 登録日 | 2010-06-03 | | 公開日 | 2010-09-22 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Computational Design of a Chain-Specific Tetracycline Repressor Heterodimer.
J.Mol.Biol., 403, 2010
|
|
2XGC
 
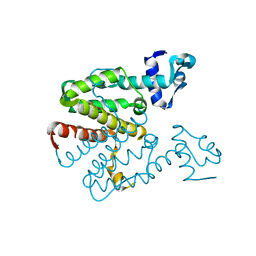 | | Crystal structure of a designed heterodimeric variant T-A(I)B of the tetracycline repressor | | 分子名称: | TETRACYCLINE REPRESSOR PROTEIN CLASS B FROM TRANSPOSON TN10, TETRACYCLINE REPRESSOR PROTEIN CLASS D | | 著者 | Stiebritz, M.T, Wengrzik, S, Richter, J.P, Muller, Y.A. | | 登録日 | 2010-06-03 | | 公開日 | 2010-09-22 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Computational Design of a Chain-Specific Tetracycline Repressor Heterodimer.
J.Mol.Biol., 403, 2010
|
|
5NL5
 
 | | Crystal structure of Zn1.7-E16V human ubiquitin (hUb) mutant adduct, from a solution 70 mM zinc acetate/1.3 mM E16V hUb | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, Polyubiquitin-B, ... | | 著者 | Fermani, S, Falini, G. | | 登録日 | 2017-04-04 | | 公開日 | 2017-04-26 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Aggregation Pathways of Native-Like Ubiquitin Promoted by Single-Point Mutation, Metal Ion Concentration, and Dielectric Constant of the Medium.
Chemistry, 24, 2018
|
|
6U4M
 
 | |
5NLF
 
 | | Crystal structure of Zn2.7-E16V human ubiquitin (hUb) mutant adduct, from a solution 100 mM zinc acetate/1.3 mM E16V hUb | | 分子名称: | ACETATE ION, Polyubiquitin-C, ZINC ION | | 著者 | Fermani, S, Falini, G. | | 登録日 | 2017-04-04 | | 公開日 | 2017-05-03 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Aggregation Pathways of Native-Like Ubiquitin Promoted by Single-Point Mutation, Metal Ion Concentration, and Dielectric Constant of the Medium.
Chemistry, 24, 2018
|
|
5NLJ
 
 | | Crystal structure of Zn3-E16V human ubiquitin (hUb) mutant adduct, from a solution 70 mM zinc acetate/20% v/v TFE/1.3 mM E16V hUb | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Fermani, S, Falini, G. | | 登録日 | 2017-04-04 | | 公開日 | 2017-04-26 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | Aggregation Pathways of Native-Like Ubiquitin Promoted by Single-Point Mutation, Metal Ion Concentration, and Dielectric Constant of the Medium.
Chemistry, 24, 2018
|
|
5NQS
 
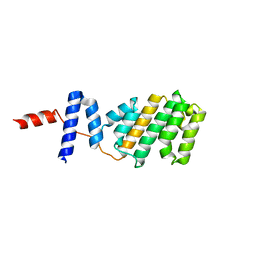 | | Structure of the Arabidopsis Thaliana TOPLESS N-terminal domain | | 分子名称: | Protein TOPLESS | | 著者 | Nanao, M.H, Arevalillo, M.R, Vinos-Poyo, T, Parcy, F, Dumas, R. | | 登録日 | 2017-04-21 | | 公開日 | 2017-07-26 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Structure of the Arabidopsis TOPLESS corepressor provides insight into the evolution of transcriptional repression.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4GMR
 
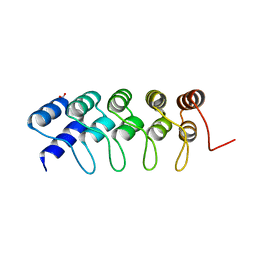 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR266. | | 分子名称: | NITRATE ION, OR266 DE NOVO PROTEIN | | 著者 | Vorobiev, S, Su, M, Parmeggiani, F, Seetharaman, J, Huang, P.-S, Mao, M, Xiao, R, Lee, D, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2012-08-16 | | 公開日 | 2012-08-29 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.377 Å) | | 主引用文献 | Computational design of self-assembling cyclic protein homo-oligomers.
NAT.CHEM., 9, 2017
|
|
3MDU
 
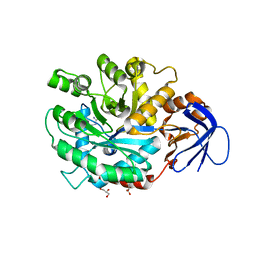 | | The structure of N-formimino-L-Glutamate Iminohydrolase from Pseudomonas aeruginosa complexed with N-Guanidino-L-Glutamate | | 分子名称: | GLYCEROL, N-carbamimidoyl-L-glutamic acid, N-formimino-L-Glutamate Iminohydrolase, ... | | 著者 | Fedorov, A.A, Fedorov, E.V, Marti-Arbona, R, Raushel, F.M, Almo, S.C. | | 登録日 | 2010-03-30 | | 公開日 | 2011-03-09 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.4003 Å) | | 主引用文献 | Structure of N-Formimino-l-glutamate Iminohydrolase from Pseudomonas aeruginosa.
Biochemistry, 54, 2015
|
|
