2RFB
 
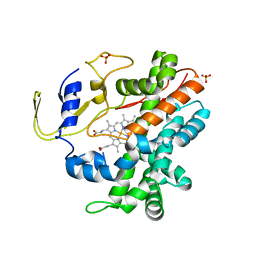 | | Crystal Structure of a Cytochrome P450 from the Thermoacidophilic Archaeon Picrophilus Torridus | | 分子名称: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | 著者 | Ho, W.W, Li, H, Poulos, T.L, Nishida, C.R, Ortiz de Montellano, P.R. | | 登録日 | 2007-09-28 | | 公開日 | 2008-01-29 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structure and Properties of CYP231A2 from the Thermoacidophilic Archaeon Picrophilus torridus.
Biochemistry, 47, 2008
|
|
8HR9
 
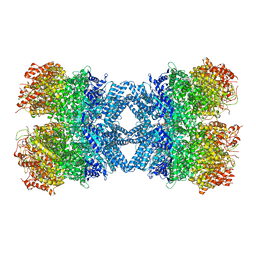 | | Structure of tetradecameric RdrA ring | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Archaeal ATPase | | 著者 | Gao, Y. | | 登録日 | 2022-12-15 | | 公開日 | 2023-02-01 | | 最終更新日 | 2023-08-16 | | 実験手法 | ELECTRON MICROSCOPY (3.03 Å) | | 主引用文献 | Molecular basis of RADAR anti-phage supramolecular assemblies.
Cell, 186, 2023
|
|
5SRI
 
 | |
5SP2
 
 | |
4WD9
 
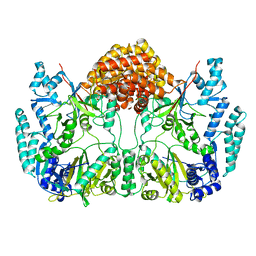 | |
8H3X
 
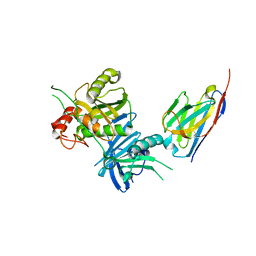 | |
4WEA
 
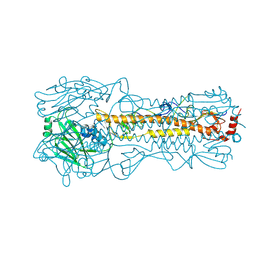 | | Structure and receptor binding prefereneces of recombinant human A(H3N2) virus hemagglutinins | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | 著者 | Yang, H, Carney, P.J, Chang, J.C, Guo, Z, Villanueva, J.M, Stevens, J. | | 登録日 | 2014-09-09 | | 公開日 | 2015-02-11 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure and receptor binding preferences of recombinant human A(H3N2) virus hemagglutinins.
Virology, 477C, 2015
|
|
5SP3
 
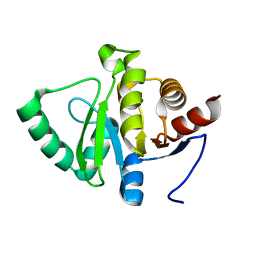 | |
8H8R
 
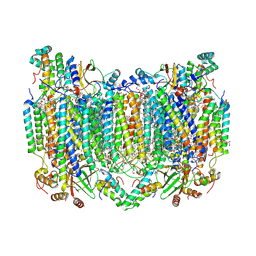 | | Bovine Heart Cytochrome c Oxidase in the Calcium-bound Fully Oxidized State | | 分子名称: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, 1,2-ETHANEDIOL, ... | | 著者 | Muramoto, K, Shinzawa-Itoh, K. | | 登録日 | 2022-10-24 | | 公開日 | 2023-02-08 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Calcium-bound structure of bovine cytochrome c oxidase.
Biochim Biophys Acta Bioenerg, 1864, 2023
|
|
5SRJ
 
 | |
4UXR
 
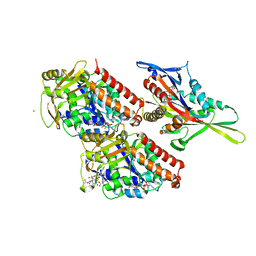 | | Conserved mechanisms of microtubule-stimulated ADP release, ATP binding, and force generation in transport kinesins | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Atherton, J, Farabella, I, Yu, I.M, Rosenfeld, S.S, Houdusse, A, Topf, M, Moores, C. | | 登録日 | 2014-08-27 | | 公開日 | 2014-09-24 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (7 Å) | | 主引用文献 | Conserved Mechanisms of Microtubule-Stimulated Adp Release, ATP Binding, and Force Generation in Transport Kinesins.
Elife, 3, 2014
|
|
8HI2
 
 | |
2RGS
 
 | | FC-fragment of monoclonal antibody IGG2B from Mus musculus | | 分子名称: | Ig gamma-2B heavy chain, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Kolenko, P, Duskova, J, Skalova, T, Dohnalek, J, Hasek, J. | | 登録日 | 2007-10-05 | | 公開日 | 2008-10-07 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | New insights into intra- and intermolecular interactions of immunoglobulins: crystal structure of mouse IgG2b-Fc at 2.1-A resolution
Immunology, 126, 2008
|
|
4WEJ
 
 | | Crystal structure of Pseudomonas aeruginosa PBP3 with a R4 substituted allyl monocarbam | | 分子名称: | (3R,4S,7Z)-7-(2-amino-1,3-thiazol-4-yl)-4-formyl-1-[({3-[(5R)-5-hydroxy-4-oxo-4,5-dihydropyridin-2-yl]-4-[3-(methylsulfonyl)propyl]-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl}sulfonyl)amino]-10,10-dimethyl-1,6-dioxo-3-(prop-2-en-1-yl)-9-oxa-2,5,8-triazaundec-7-en-11-oic acid, Penicillin-binding protein 3 | | 著者 | Ferguson, A.D. | | 登録日 | 2014-09-10 | | 公開日 | 2015-04-22 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.045 Å) | | 主引用文献 | SAR and Structural Analysis of Siderophore-Conjugated Monocarbam Inhibitors of Pseudomonas aeruginosa PBP3.
Acs Med.Chem.Lett., 6, 2015
|
|
2RSO
 
 | |
5SRK
 
 | |
8HRA
 
 | | Structure of heptameric RdrA ring in RNA-loading state | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Archaeal ATPase, RNA (5'-R(P*GP*UP*CP*CP*AP*GP*CP*GP*UP*CP*AP*UP*CP*GP*CP*UP*GP*GP*AP*C)-3') | | 著者 | Gao, Y. | | 登録日 | 2022-12-15 | | 公開日 | 2023-02-01 | | 最終更新日 | 2023-08-16 | | 実験手法 | ELECTRON MICROSCOPY (3.76 Å) | | 主引用文献 | Molecular basis of RADAR anti-phage supramolecular assemblies.
Cell, 186, 2023
|
|
2ROT
 
 | | Structure of chimeric variant of SH3 domain- SHH | | 分子名称: | Spectrin alpha chain, brain | | 著者 | Kutyshenko, N.P, Prokhorov, D.A, Timchenko, M.A, Kudrevatykh, Y.A, Gushchina, L.V, Khristoforov, V.S, Filimonov, V.V. | | 登録日 | 2008-04-10 | | 公開日 | 2009-04-28 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure and dynamics of the chimeric SH3 domains, SHH- and SHA-"Bergeracs".
Biochim.Biophys.Acta, 1794, 2009
|
|
4URP
 
 | | The Crystal structure of Nitroreductase from Saccharomyces cerevisiae | | 分子名称: | FATTY ACID REPRESSION MUTANT PROTEIN 2 | | 著者 | Song, H.-N, Woo, E.-J, Bang, S.-Y, Jung, D.-G, Park, S.-G. | | 登録日 | 2014-07-01 | | 公開日 | 2015-04-29 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.991 Å) | | 主引用文献 | Crystal Structure of the Fungal Nitroreductase Frm2 from Saccharomyces Cerevisiae.
Protein Sci., 24, 2015
|
|
8H9A
 
 | | Crystal structure of chemically modified E. coli ThrS catalytic domain 2 | | 分子名称: | N-(2,3-dihydroxybenzoyl)-4-(4-nitrophenyl)-L-threonine, Threonine--tRNA ligase, ZINC ION | | 著者 | Qiao, H, Xia, M, Wang, J, Fang, P. | | 登録日 | 2022-10-25 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Tyrosine-targeted covalent inhibition of a tRNA synthetase aided by zinc ion.
Commun Biol, 6, 2023
|
|
5SRL
 
 | |
5SP0
 
 | |
4V05
 
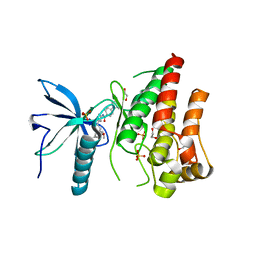 | | FGFR1 in complex with AZD4547. | | 分子名称: | 1,2-ETHANEDIOL, FIBROBLAST GROWTH FACTOR RECEPTOR 1 (FMS-RELATED TYROSINE KINASE 2, PFEIFFER SYNDROME), ... | | 著者 | Tucker, J, Klein, T, Breed, J, Breeze, A, Overman, R, Phillips, C, Norman, R.A. | | 登録日 | 2014-09-10 | | 公開日 | 2014-12-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.57 Å) | | 主引用文献 | Structural Insights Into Fgfr Kinase Isoform Selectivity: Diverse Binding Modes of Azd4547 and Ponatinib in Complex with Fgfr1 and Fgfr4
Structure, 22, 2014
|
|
8H9D
 
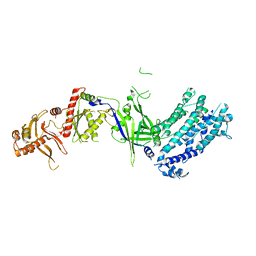 | | Crystal structure of Cas12a protein | | 分子名称: | Cas12A, MAGNESIUM ION, RNA (5'-R(P*AP*AP*UP*UP*UP*CP*UP*AP*CP*UP*AP*AP*GP*UP*GP*UP*AP*GP*AP*UP*C)-3'), ... | | 著者 | Jianwei, L, Jobichen, C, Sivaraman, J. | | 登録日 | 2022-10-25 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structures of apo Cas12a and its complex with crRNA and DNA reveal the dynamics of ternary complex formation and target DNA cleavage.
Plos Biol., 21, 2023
|
|
2RU7
 
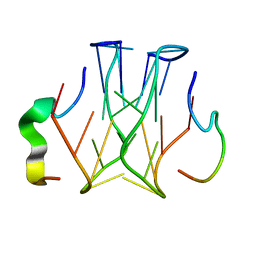 | | Refined structure of RNA aptamer in complex with the partial binding peptide of prion protein | | 分子名称: | P16 peptide from Major prion protein, RNA_(5'-R(*GP*GP*AP*GP*GP*AP*GP*GP*AP*GP*GP*A)-3') | | 著者 | Hayashi, T, Oshima, H, Mashima, T, Nagata, T, Katahira, M, Kinoshita, M. | | 登録日 | 2013-12-24 | | 公開日 | 2014-05-21 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Binding of an RNA aptamer and a partial peptide of a prion protein: crucial importance of water entropy in molecular recognition.
Nucleic Acids Res., 42, 2014
|
|
