3NFF
 
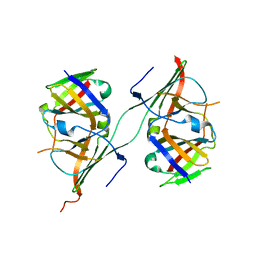 | | Crystal structure of extended Dimerization module of RNA polymerase I subcomplex A49/A34.5 | | 分子名称: | RNA polymerase I subunit A34.5, RNA polymerase I subunit A49 | | 著者 | Geiger, S.R, Lorenzen, K, Schreieck, A, Hanecker, P, Kostrewa, D, Heck, A.J.R, Cramer, P. | | 登録日 | 2010-06-10 | | 公開日 | 2010-09-08 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.24 Å) | | 主引用文献 | RNA Polymerase I Contains a TFIIF-Related DNA-Binding Subcomplex.
Mol.Cell, 39, 2010
|
|
6KML
 
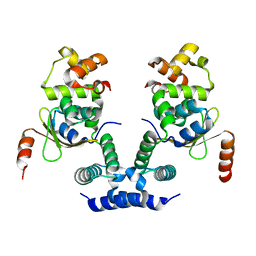 | | 2.09 Angstrom resolution crystal structure of tetrameric HigBA toxin-antitoxin complex from E.coli | | 分子名称: | Antitoxin HigA, mRNA interferase toxin HigB | | 著者 | Jadhav, P, Sinha, V.K, Rothweiler, U, Singh, M. | | 登録日 | 2019-07-31 | | 公開日 | 2020-11-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.095 Å) | | 主引用文献 | 2.09 angstrom Resolution structure of E. coli HigBA toxin-antitoxin complex reveals an ordered DNA-binding domain and intrinsic dynamics in antitoxin.
Biochem.J., 477, 2020
|
|
6KMQ
 
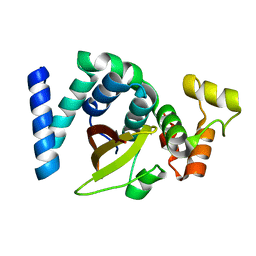 | | 2.3 Angstrom resolution structure of dimeric HigBA toxin-antitoxin complex from E. coli | | 分子名称: | Antitoxin HigA, mRNA interferase toxin HigB | | 著者 | Jadhav, P, Sinha, V.K, Rothweiler, U, Singh, M. | | 登録日 | 2019-07-31 | | 公開日 | 2020-11-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | 2.09 angstrom Resolution structure of E. coli HigBA toxin-antitoxin complex reveals an ordered DNA-binding domain and intrinsic dynamics in antitoxin.
Biochem.J., 477, 2020
|
|
4ZPK
 
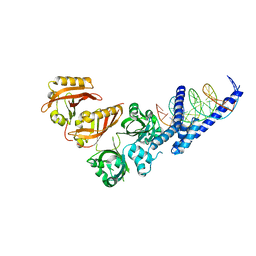 | | Crystal Structure of the Heterodimeric HIF-2a:ARNT Complex with HRE DNA | | 分子名称: | Aryl hydrocarbon receptor nuclear translocator, DNA (5'-D(*CP*AP*CP*GP*AP*CP*CP*CP*GP*CP*AP*CP*GP*TP*AP*CP*GP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*GP*CP*TP*GP*CP*GP*TP*AP*CP*GP*TP*GP*CP*GP*GP*GP*TP*CP*GP*T)-3'), ... | | 著者 | Wu, D, Potluri, N, Lu, J, Kim, Y, Rastinejad, F. | | 登録日 | 2015-05-08 | | 公開日 | 2015-08-12 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Structural integration in hypoxia-inducible factors.
Nature, 524, 2015
|
|
4ZPR
 
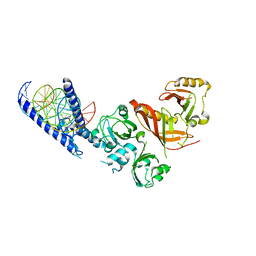 | | Crystal Structure of the Heterodimeric HIF-1a:ARNT Complex with HRE DNA | | 分子名称: | Aryl hydrocarbon receptor nuclear translocator, DNA (5'-D(*CP*AP*CP*GP*AP*CP*CP*CP*GP*CP*AP*CP*GP*TP*AP*CP*GP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*GP*CP*TP*GP*CP*GP*TP*AP*CP*GP*TP*GP*CP*GP*GP*GP*TP*CP*GP*T)-3'), ... | | 著者 | Wu, D, Potluri, N, Lu, J, Kim, Y, Rastinejad, F. | | 登録日 | 2015-05-08 | | 公開日 | 2015-08-12 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.902 Å) | | 主引用文献 | Structural integration in hypoxia-inducible factors.
Nature, 524, 2015
|
|
6M5S
 
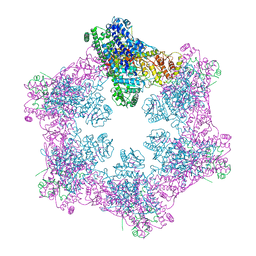 | | The coordinates of the apo hexameric terminase complex | | 分子名称: | Tripartite terminase subunit 1, Tripartite terminase subunit 2, Tripartite terminase subunit 3, ... | | 著者 | Yang, Y.X, Yang, P, Wang, N, Chen, Z.H, Zhou, Z.H, Rao, Z.H, Wang, X.X. | | 登録日 | 2020-03-11 | | 公開日 | 2020-10-28 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Architecture of the herpesvirus genome-packaging complex and implications for DNA translocation.
Protein Cell, 11, 2020
|
|
6M5U
 
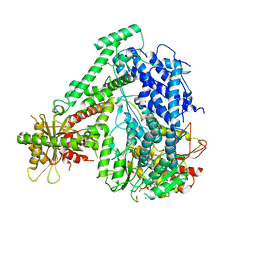 | | The coordinates of the monomeric terminase complex in the presence of the ADP-BeF3 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | 著者 | Yang, Y.X, Yang, P, Wang, N, Zhu, L, Zhou, Z.H, Rao, Z.H, Wang, X.X. | | 登録日 | 2020-03-11 | | 公開日 | 2020-10-28 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Architecture of the herpesvirus genome-packaging complex and implications for DNA translocation.
Protein Cell, 11, 2020
|
|
6O47
 
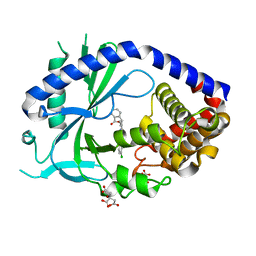 | | human cGAS core domain (K427E/K428E) bound with RU-521 | | 分子名称: | (3~{S})-3-[1-[4,5-bis(chloranyl)-1~{H}-benzimidazol-2-yl]-3-methyl-5-oxidanyl-pyrazol-4-yl]-3~{H}-2-benzofuran-1-one, 2-(4,5-dichloro-1H-benzimidazol-2-yl)-5-methyl-4-[(1R)-3-oxo-1,3-dihydro-2-benzofuran-1-yl]-1,2-dihydro-3H-pyrazol-3-one, CITRIC ACID, ... | | 著者 | Xie, W, Lama, L, Adura, C, Glickman, J.F, Tuschl, T, Patel, D.J. | | 登録日 | 2019-02-28 | | 公開日 | 2019-05-29 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.196 Å) | | 主引用文献 | Human cGAS catalytic domain has an additional DNA-binding interface that enhances enzymatic activity and liquid-phase condensation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6M5R
 
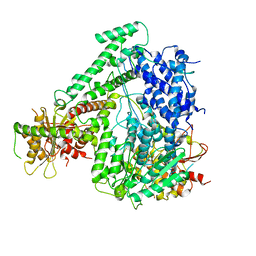 | | The coordinates of the apo monomeric terminase complex | | 分子名称: | Tripartite terminase subunit 1, Tripartite terminase subunit 2, Tripartite terminase subunit 3, ... | | 著者 | Yang, Y.X, Yang, P, Wang, N, Chen, Z.H, Zhou, Z.H, Rao, Z.H, Wang, X.X. | | 登録日 | 2020-03-11 | | 公開日 | 2020-10-28 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Architecture of the herpesvirus genome-packaging complex and implications for DNA translocation.
Protein Cell, 11, 2020
|
|
6M5V
 
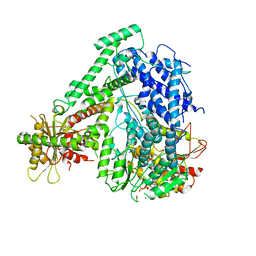 | | The coordinate of the hexameric terminase complex in the presence of the ADP-BeF3 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | 著者 | Yang, Y.X, Yang, P, Wang, N, Chen, Z.H, Zhou, Z.H, Rao, Z.H, Wang, X.X. | | 登録日 | 2020-03-11 | | 公開日 | 2020-10-28 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Architecture of the herpesvirus genome-packaging complex and implications for DNA translocation.
Protein Cell, 11, 2020
|
|
4XR1
 
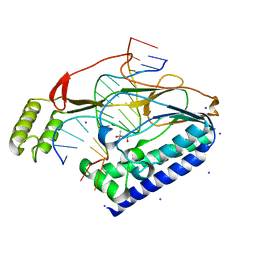 | |
1IDY
 
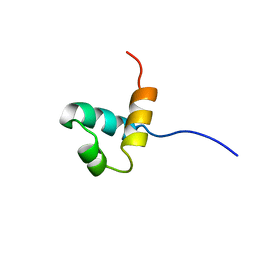 | |
4XR2
 
 | |
4XR3
 
 | |
7T5V
 
 | |
7T5W
 
 | |
6PQX
 
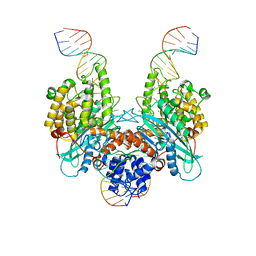 | | Cryo-EM structure of HzTransib/nicked TIR substrate DNA hairpin forming complex (HFC) | | 分子名称: | CALCIUM ION, DNA (5'-D(P*CP*AP*CP*GP*GP*TP*GP*GP*AP*TP*CP*GP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*CP*TP*GP*GP*CP*CP*TP*AP*GP*AP*TP*CP*T)-3'), ... | | 著者 | Liu, C, Yang, Y, Schatz, D.G. | | 登録日 | 2019-07-10 | | 公開日 | 2019-10-09 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Structures of a RAG-like transposase during cut-and-paste transposition.
Nature, 575, 2019
|
|
4L85
 
 | |
7TMY
 
 | |
6NE0
 
 | | Structure of double-stranded target DNA engaged Csy complex from Pseudomonas aeruginosa (PA-14) | | 分子名称: | CRISPR RNA (60-MER), CRISPR target DNA (44-MER), CRISPR-associated endonuclease Cas6/Csy4, ... | | 著者 | Chowdhury, S, Rollins, M.F, Carter, J, Golden, S.M, Miettinen, H.M, Santiago-Frangos, A, Faith, D, Lawrence, M.C, Wiedenheft, B, Lander, G.C. | | 登録日 | 2018-12-15 | | 公開日 | 2018-12-26 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure Reveals a Mechanism of CRISPR-RNA-Guided Nuclease Recruitment and Anti-CRISPR Viral Mimicry.
Mol. Cell, 74, 2019
|
|
7MR1
 
 | |
7MR0
 
 | |
7MR2
 
 | |
1CI6
 
 | | TRANSCRIPTION FACTOR ATF4-C/EBP BETA BZIP HETERODIMER | | 分子名称: | BETA-MERCAPTOETHANOL, FE (III) ION, TRANSCRIPTION FACTOR ATF-4, ... | | 著者 | Podust, L.M, Kim, Y. | | 登録日 | 1999-04-07 | | 公開日 | 2000-12-04 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of the CCAAT box/enhancer-binding protein beta activating transcription factor-4 basic leucine zipper heterodimer in the absence of DNA
J.Biol.Chem., 276, 2001
|
|
1IDZ
 
 | |
