5VNE
 
 | |
4P0J
 
 | |
5VNI
 
 | |
5Q0T
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | 2-phenyl-N-(propan-2-yl)-6-[(thiophen-2-yl)sulfonyl]-4,5,6,7-tetrahydro-1H-pyrrolo[2,3-c]pyridine-1-carboxamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q11
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3, N,N-dicyclohexyl-3-(2,4-dichlorophenyl)-5-methyl-1,2-oxazole-4-carboxamide | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q18
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | (2S)-2-cyclohexyl-2-{5,6-difluoro-2-[(R)-methoxy(phenyl)methyl]-1H-benzimidazol-1-yl}-N-(trans-4-hydroxycyclohexyl)acetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
4OIF
 
 | | 3D structure of Gan42B, a GH42 beta-galactosidase from G. | | 分子名称: | Beta-galactosidase, GLYCEROL, ZINC ION | | 著者 | Solomon, H.V, Tabachnikov, O, Feinberg, H, Shoham, Y, Shoham, G. | | 登録日 | 2014-01-19 | | 公開日 | 2015-02-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.448 Å) | | 主引用文献 | Crystallization and preliminary crystallographic analysis of GanB, a GH42 intracellular beta-galactosidase from Geobacillus stearothermophilus.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4O9H
 
 | | Structure of Interleukin-6 in complex with a Camelid Fab fragment | | 分子名称: | Heavy Chain of the Camelid Fab fragment 61H7, Interleukin-6, Light Chain of the Camelid Fab fragment 61H7 | | 著者 | Klarenbeek, A, Blanchetot, C, Schragel, G, Sadi, A.S, Ongenae, N, Hemrika, W, Wijdenes, J, Spinelli, S, Desmyter, A, Cambillau, C, Hultberg, A, Kretz-rommel, A, Dreier, T, De haard, H.J.W, Roovers, R.C. | | 登録日 | 2014-01-02 | | 公開日 | 2015-04-15 | | 最終更新日 | 2015-04-29 | | 実験手法 | X-RAY DIFFRACTION (2.42 Å) | | 主引用文献 | Combining residues of naturally-occurring Camelid somatic affinity variants yields ultra-potent human therapeutic IL-6 antibodies
To be Published
|
|
4OGC
 
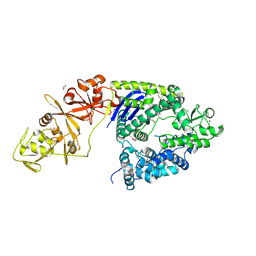 | | Crystal structure of the Type II-C Cas9 enzyme from Actinomyces naeslundii | | 分子名称: | ACETATE ION, HNH endonuclease domain protein, MAGNESIUM ION, ... | | 著者 | Jiang, F, Ma, E, Lin, S, Doudna, J.A. | | 登録日 | 2014-01-15 | | 公開日 | 2014-02-12 | | 最終更新日 | 2014-03-26 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structures of Cas9 endonucleases reveal RNA-mediated conformational activation.
Science, 343, 2014
|
|
4P0L
 
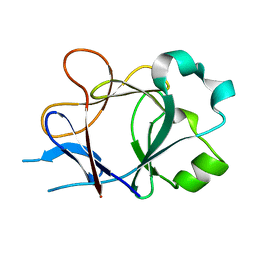 | |
4O8Q
 
 | | Crystal structure of bovine MHD domain of the COPI delta subunit at 2.15 A resolution | | 分子名称: | Coatomer subunit delta, FORMIC ACID | | 著者 | Lahav, A, Rozenberg, H, Cassel, D, Adir, N. | | 登録日 | 2013-12-29 | | 公開日 | 2015-01-07 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structure of the bovine COPI delta subunit mu homology domain at 2.15 angstrom resolution.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7JTI
 
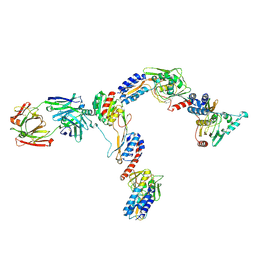 | | Interphotoreceptor retinoid-binding protein (IRBP) in complex with a monoclonal antibody (F3F5 mAb5) | | 分子名称: | Retinol-binding protein 3, mAb5 Fab heavy chain, mAb5 Fab light chain | | 著者 | Sears, A.E, Albiez, S, Gulati, S, Wang, B, Kiser, P, Kovacik, L, Engel, A, Stahlberg, H, Palczewski, K. | | 登録日 | 2020-08-17 | | 公開日 | 2020-10-07 | | 最終更新日 | 2020-12-16 | | 実験手法 | ELECTRON MICROSCOPY (7.4 Å) | | 主引用文献 | Single particle cryo-EM of the complex between interphotoreceptor retinoid-binding protein and a monoclonal antibody.
Faseb J., 34, 2020
|
|
7JU4
 
 | | Radial spoke 2 stalk, IDAc, and N-DRC attached with doublet microtubule | | 分子名称: | 28 kDa inner dynein arm light chain, axonemal, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Gui, M, Ma, M, Sze-Tu, E, Wang, X, Koh, F, Zhong, E, Berger, B, Davis, J, Dutcher, S, Zhang, R, Brown, A. | | 登録日 | 2020-08-19 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structures of radial spokes and associated complexes important for ciliary motility.
Nat.Struct.Mol.Biol., 28, 2021
|
|
4N1L
 
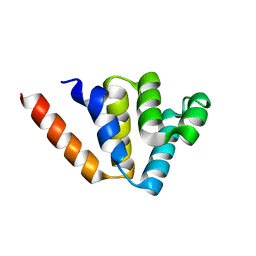 | | Crystal structures of NLRP14 pyrin domain reveal a conformational switch mechanism, regulating its molecular interactions | | 分子名称: | NACHT, LRR and PYD domains-containing protein 14 | | 著者 | Eibl, C, Hessenberger, M, Wenger, J, Brandstetter, H. | | 登録日 | 2013-10-04 | | 公開日 | 2014-07-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.986 Å) | | 主引用文献 | Structures of the NLRP14 pyrin domain reveal a conformational switch mechanism regulating its molecular interactions.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4N3A
 
 | |
7JTK
 
 | | Radial spoke 1 isolated from Chlamydomonas reinhardtii | | 分子名称: | Cytochrome b5 heme-binding domain-containing protein, Dynein 8 kDa light chain, flagellar outer arm, ... | | 著者 | Gui, M, Ma, M, Sze-Tu, E, Wang, X, Koh, F, Zhong, E, Berger, B, Davis, J, Dutcher, S, Zhang, R, Brown, A. | | 登録日 | 2020-08-17 | | 公開日 | 2020-12-16 | | 最終更新日 | 2021-01-27 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structures of radial spokes and associated complexes important for ciliary motility.
Nat.Struct.Mol.Biol., 28, 2021
|
|
4N3C
 
 | |
4N42
 
 | | Crystal structure of allergen protein scam1 from Scadoxus multiflorus | | 分子名称: | PHOSPHATE ION, Xylanase and alpha-amylase inhibitor protein isoform III | | 著者 | Singh, A, Kumar, S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | 登録日 | 2013-10-08 | | 公開日 | 2013-10-30 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of allergen protein scam1 from Scadoxus multiflorus
To be published
|
|
5Q1E
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | 5-bromo-1-{[4-(1H-tetrazol-5-yl)phenyl]methyl}-1'-(thiophene-2-sulfonyl)spiro[indole-3,4'-piperidin]-2(1H)-one, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
4N1K
 
 | | Crystal structures of NLRP14 pyrin domain reveal a conformational switch mechanism, regulating its molecular interactions | | 分子名称: | NACHT, LRR and PYD domains-containing protein 14 | | 著者 | Eibl, C, Hessenberger, M, Wenger, J, Brandstetter, H. | | 登録日 | 2013-10-04 | | 公開日 | 2014-07-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structures of the NLRP14 pyrin domain reveal a conformational switch mechanism regulating its molecular interactions.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OJP
 
 | |
5VNO
 
 | | Crystal structure of Sec23a/Sec24a/Sec22 | | 分子名称: | Protein transport protein Sec23A, Protein transport protein Sec24A, Vesicle-trafficking protein SEC22b, ... | | 著者 | Ma, W, Goldberg, J. | | 登録日 | 2017-05-01 | | 公開日 | 2017-07-05 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.905 Å) | | 主引用文献 | ER retention is imposed by COPII protein sorting and attenuated by 4-phenylbutyrate.
Elife, 6, 2017
|
|
4OUU
 
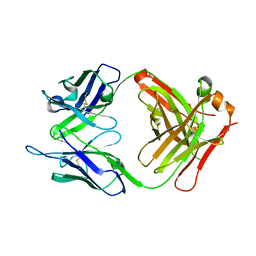 | | anti-MT1-MMP monoclonal antibody | | 分子名称: | anti_MT1-MMP Heavy chain, anti_MT1-MMP light chain | | 著者 | Udi, Y, Grossman, M, Solomonov, I, Dym, O, Rozenberg, H, Koziol, A, Cuniasse, P, Dive, V, Arroyo, A.G, Irit, S, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2014-02-19 | | 公開日 | 2014-12-17 | | 最終更新日 | 2015-01-28 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Inhibition mechanism of membrane metalloprotease by an exosite-swiveling conformational antibody.
Structure, 23, 2015
|
|
4OBA
 
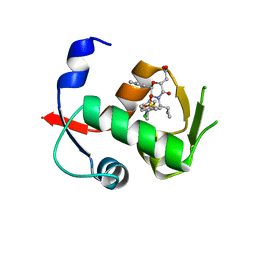 | | Co-crystal structure of MDM2 with Inhibitor Compound 4 | | 分子名称: | E3 ubiquitin-protein ligase Mdm2, [(2R,5R,6R)-4-[(1S)-2-(tert-butylsulfonyl)-1-cyclopropylethyl]-6-(3-chlorophenyl)-5-(4-chlorophenyl)-3-oxomorpholin-2-yl]acetic acid | | 著者 | Shaffer, P.L, Huang, X, Yakowec, P, Long, A.M. | | 登録日 | 2014-01-07 | | 公開日 | 2014-03-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.602 Å) | | 主引用文献 | Selective and Potent Morpholinone Inhibitors of the MDM2-p53 Protein-Protein Interaction.
J.Med.Chem., 57, 2014
|
|
5Q0L
 
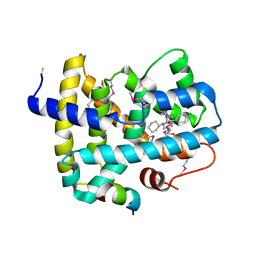 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | (2S)-N,2-dicyclohexyl-2-{2-[4-(hydroxymethyl)phenyl]-1H-benzimidazol-1-yl}acetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
