1J0H
 
 | |
1J0K
 
 | |
1J0I
 
 | |
7VPY
 
 | | Crystal structure of the neutralizing nanobody P86 against SARS-CoV-2 | | 分子名称: | 1,2-ETHANEDIOL, Nanobody, SULFATE ION | | 著者 | Maeda, R, Fujita, J, Konishi, Y, Kazuma, Y, Yamazaki, H, Anzai, I, Yamaguchi, K, Kasai, K, Nagata, K, Yamaoka, Y, Miyakawa, K, Ryo, A, Shirakawa, K, Makino, F, Matsuura, Y, Inoue, T, Imura, A, Namba, K, Takaori-Kondo, A. | | 登録日 | 2021-10-18 | | 公開日 | 2022-07-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | A panel of nanobodies recognizing conserved hidden clefts of all SARS-CoV-2 spike variants including Omicron.
Commun Biol, 5, 2022
|
|
7VQ0
 
 | | Cryo-EM structure of the SARS-CoV-2 spike protein (2-up RBD) bound to neutralizing nanobodies P86 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Maeda, R, Fujita, J, Konishi, Y, Kazuma, Y, Yamazaki, H, Anzai, I, Yamaguchi, K, Kasai, K, Nagata, K, Yamaoka, Y, Miyakawa, K, Ryo, A, Shirakawa, K, Makino, F, Matsuura, Y, Inoue, T, Imura, A, Namba, K, Takaori-Kondo, A. | | 登録日 | 2021-10-18 | | 公開日 | 2022-07-20 | | 実験手法 | ELECTRON MICROSCOPY (3.03 Å) | | 主引用文献 | A panel of nanobodies recognizing conserved hidden clefts of all SARS-CoV-2 spike variants including Omicron.
Commun Biol, 5, 2022
|
|
1IV8
 
 | |
5H2X
 
 | |
5H2W
 
 | |
5GUA
 
 | | Structure of biotin carboxyl carrier protein from pyrococcus horikoshi OT3 (delta N79) A138Y mutant | | 分子名称: | 149aa long hypothetical methylmalonyl-CoA decarboxylase gamma chain | | 著者 | Yamada, K, Kunishima, N, Matsuura, Y, Nakai, K, Naitow, H, Fukasawa, Y, Tomii, K. | | 登録日 | 2016-08-26 | | 公開日 | 2017-08-30 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Designing better diffracting crystals of biotin carboxyl carrier protein from Pyrococcus horikoshii by a mutation based on the crystal-packing propensity of amino acids.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5GU9
 
 | | Structure of biotin carboxyl carrier protein from pyrococcus horikoshi OT3 (delta N79) A138I mutant | | 分子名称: | 149aa long hypothetical methylmalonyl-CoA decarboxylase gamma chain | | 著者 | Yamada, K, Kunishima, N, Matsuura, Y, Nakai, K, Naitow, H, Fukasawa, Y, Tomii, K. | | 登録日 | 2016-08-26 | | 公開日 | 2017-08-30 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Designing better diffracting crystals of biotin carboxyl carrier protein from Pyrococcus horikoshii by a mutation based on the crystal-packing propensity of amino acids.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5H2V
 
 | |
5GU8
 
 | | Structure of biotin carboxyl carrier protein from pyrococcus horikoshi OT3 (delta N79) wild type | | 分子名称: | 149aa long hypothetical methylmalonyl-CoA decarboxylase gamma chain, SODIUM ION | | 著者 | Yamada, K, Kunishima, N, Matsuura, Y, Nakai, K, Naitow, H, Fukasawa, Y, Tomii, K. | | 登録日 | 2016-08-26 | | 公開日 | 2017-08-30 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Designing better diffracting crystals of biotin carboxyl carrier protein from Pyrococcus horikoshii by a mutation based on the crystal-packing propensity of amino acids.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
7E7F
 
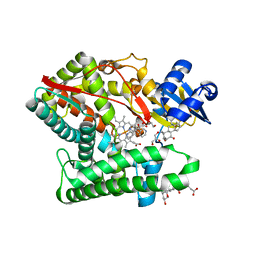 | | Human CYP11B1 mutant in complex with metyrapone | | 分子名称: | CHOLIC ACID, Cytochrome P450 11B1, mitochondrial, ... | | 著者 | Mukai, K, Sugimoto, H, Reiko, S, Matsuura, T, Hishiki, T, Kagawa, N. | | 登録日 | 2021-02-26 | | 公開日 | 2022-01-05 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Spatially restricted substrate-binding site of cortisol-synthesizing CYP11B1 limits multiple hydroxylations and hinders aldosterone synthesis.
Curr Res Struct Biol, 3, 2021
|
|
5XVB
 
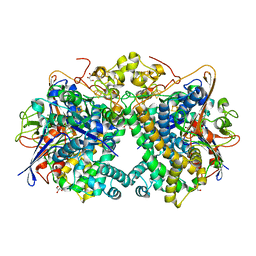 | | [NiFe]-hydrogenase (Hyb-type) from Citrobacter sp. S-77 in an H2-reduced condition | | 分子名称: | FE3-S4 CLUSTER, GLYCEROL, IRON/SULFUR CLUSTER, ... | | 著者 | Nishikawa, K, Matsuura, H, Muhd Noor, N.D, Tai, H, Hirota, S, Kim, J, Kang, J, Tateno, M, Yoon, K.S, Ogo, S, Shomura, Y, Higuchi, Y. | | 登録日 | 2017-06-27 | | 公開日 | 2018-06-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Redox-dependent conformational changes of a proximal [4Fe-4S] cluster in Hyb-type [NiFe]-hydrogenase to protect the active site from O2.
Chem.Commun.(Camb.), 54, 2018
|
|
5XVD
 
 | | [NiFe]-hydrogenase (Hyb-type) from Citrobacter sp. S-77 in an air-oxidized condition | | 分子名称: | FE3-S4 CLUSTER, FE4-S4-O CLUSTER, GLYCEROL, ... | | 著者 | Nishikawa, K, Matsuura, H, Muhd Noor, N.D, Tai, H, Hirota, S, Kim, J, Kang, J, Tateno, M, Yoon, K.S, Ogo, S, Shomura, Y, Higuchi, Y. | | 登録日 | 2017-06-27 | | 公開日 | 2018-06-27 | | 最終更新日 | 2019-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Redox-dependent conformational changes of a proximal [4Fe-4S] cluster in Hyb-type [NiFe]-hydrogenase to protect the active site from O2.
Chem.Commun.(Camb.), 54, 2018
|
|
5XZX
 
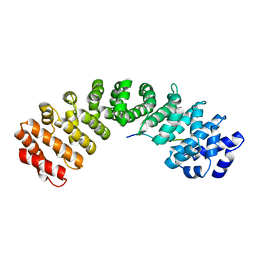 | |
5YU4
 
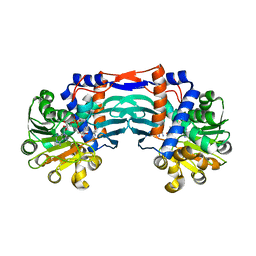 | | Structural basis for recognition of L-lysine, L-ornithine, and L-2,4-diamino butyric acid by lysine cyclodeaminase | | 分子名称: | 2,4-DIAMINOBUTYRIC ACID, Lysine cyclodeaminase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Min, K.J, Yoon, H.J, Matsuura, A, Kim, Y.H, Lee, H.H. | | 登録日 | 2017-11-20 | | 公開日 | 2018-05-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.144 Å) | | 主引用文献 | Structural Basis for Recognition of L-lysine, L-ornithine, and L-2,4-diamino Butyric Acid by Lysine Cyclodeaminase.
Mol. Cells, 41, 2018
|
|
5XVC
 
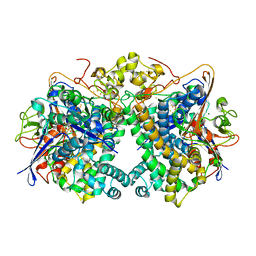 | | [NiFe]-hydrogenase (Hyb-type) from Citrobacter sp. S-77 in a ferricyanide-oxidized condition | | 分子名称: | DI(HYDROXYETHYL)ETHER, FE3-S4 CLUSTER, FE4-S4-O CLUSTER, ... | | 著者 | Nishikawa, K, Matsuura, H, Muhd Noor, N.D, Tai, H, Hirota, S, Kim, J, Kang, J, Tateno, M, Yoon, K.S, Ogo, S, Shomura, Y, Higuchi, Y. | | 登録日 | 2017-06-27 | | 公開日 | 2018-06-27 | | 最終更新日 | 2019-07-10 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Redox-dependent conformational changes of a proximal [4Fe-4S] cluster in Hyb-type [NiFe]-hydrogenase to protect the active site from O2.
Chem.Commun.(Camb.), 54, 2018
|
|
5XW4
 
 | |
5YU3
 
 | | Structural basis for recognition of L-lysine, L-ornithine, and L-2,4-diamino butyric acid by lysine cyclodeaminase | | 分子名称: | Lysine cyclodeaminase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROLINE, ... | | 著者 | Min, K.J, Yoon, H.J, Matsuura, A, Kim, Y.H, Lee, H.H. | | 登録日 | 2017-11-20 | | 公開日 | 2018-05-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Structural Basis for Recognition of L-lysine, L-ornithine, and L-2,4-diamino Butyric Acid by Lysine Cyclodeaminase.
Mol. Cells, 41, 2018
|
|
5YU1
 
 | | Structural basis for recognition of L-lysine, L-ornithine, and L-2,4-diamino butyric acid by lysine cyclodeaminase | | 分子名称: | (2S)-piperidine-2-carboxylic acid, Lysine cyclodeaminase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Min, K.J, Yoon, H.J, Matsuura, A, Kim, Y.H, Lee, H.H. | | 登録日 | 2017-11-20 | | 公開日 | 2018-05-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.923 Å) | | 主引用文献 | Structural Basis for Recognition of L-lysine, L-ornithine, and L-2,4-diamino Butyric Acid by Lysine Cyclodeaminase.
Mol. Cells, 41, 2018
|
|
5XOJ
 
 | | Crystal structure of Xpo1p-PKI-Nup42p-Gsp1p-GTP complex | | 分子名称: | Exportin-1, GTP-binding nuclear protein, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Koyama, M, Shirai, N, Matsuura, Y. | | 登録日 | 2017-05-29 | | 公開日 | 2017-08-16 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of the Xpo1p nuclear export complex bound to the SxFG/PxFG repeats of the nucleoporin Nup42p
Genes Cells, 22, 2017
|
|
5XW5
 
 | |
5X8N
 
 | |
5ZGO
 
 | |
