8FPR
 
 | |
1NTB
 
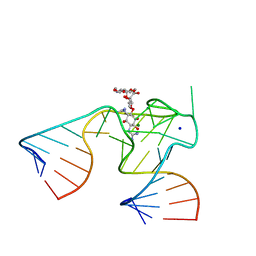 | | 2.9 A crystal structure of Streptomycin RNA-aptamer complex | | 分子名称: | 5'-R(*CP*GP*GP*CP*AP*CP*CP*AP*CP*GP*GP*UP*CP*GP*GP*AP*UP*C)-3', 5'-R(*GP*GP*AP*UP*CP*GP*CP*AP*UP*UP*UP*GP*GP*AP*CP*UP*UP*CP*UP*GP*CP*C)-3', MAGNESIUM ION, ... | | 著者 | Tereshko, V, Skripkin, E, Patel, D.J. | | 登録日 | 2003-01-29 | | 公開日 | 2003-05-13 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Encapsulating Streptomycin within a small 40-mer RNA
CHEM.BIOL., 10, 2003
|
|
8FPU
 
 | |
8FRY
 
 | |
8OM8
 
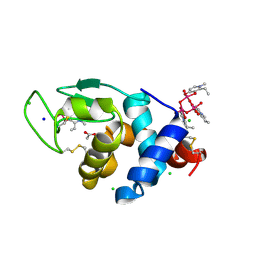 | | X-ray structure of lysozyme obtained upon reaction with [VIVO(empp)2] (Structure A) | | 分子名称: | 1-methyl-2-ethyl-3-hydroxy-4(1H)-pyridinone)V(IV)O4, ACETATE ION, CHLORIDE ION, ... | | 著者 | Paolillo, M, Ferraro, G, Merlino, A. | | 登録日 | 2023-03-31 | | 公開日 | 2023-06-07 | | 実験手法 | X-RAY DIFFRACTION (1.08 Å) | | 主引用文献 | Implications of Protein Interaction in the Speciation of Potential V IV O-Pyridinone Drugs.
Inorg.Chem., 62, 2023
|
|
6O3C
 
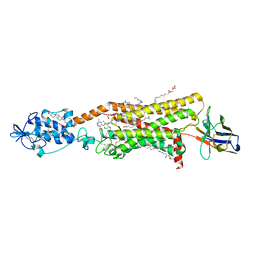 | | Crystal structure of active Smoothened bound to SAG21k, cholesterol, and NbSmo8 | | 分子名称: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-chloro-4,7-difluoro-N-{[2-methoxy-5-(pyridin-4-yl)phenyl]methyl}-N-[trans-4-(methylamino)cyclohexyl]-1-benzothiophene-2-carboxamide, ... | | 著者 | Deshpande, I.S, Liang, J, Hedeen, D, Roberts, K.J, Zhang, Y, Ha, B, Latorraca, N.R, Faust, B, Dror, R.O, Beachy, P.A, Myers, B.R, Manglik, A. | | 登録日 | 2019-02-26 | | 公開日 | 2019-07-03 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Smoothened stimulation by membrane sterols drives Hedgehog pathway activity.
Nature, 571, 2019
|
|
8OMT
 
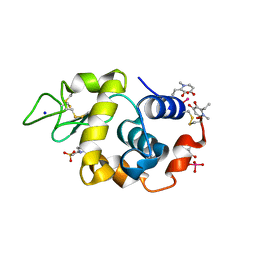 | | X-ray structure of lysozyme obtained upon reaction with [VIVO(empp)2] (Structure C) | | 分子名称: | 1-methyl-2-ethyl-3-hydroxy-4(1H)-pyridinone)V(IV)O4, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Lysozyme C, ... | | 著者 | Paolillo, M, Merlino, A, Ferraro, G. | | 登録日 | 2023-03-31 | | 公開日 | 2023-06-07 | | 実験手法 | X-RAY DIFFRACTION (1.097 Å) | | 主引用文献 | Implications of Protein Interaction in the Speciation of Potential V IV O-Pyridinone Drugs.
Inorg.Chem., 62, 2023
|
|
8OEP
 
 | | Crystal structure of the PTPN3 PDZ domain bound to the HPV18 E6 oncoprotein C-terminal peptide | | 分子名称: | Protein E6, SODIUM ION, Tyrosine-protein phosphatase non-receptor type 3 | | 著者 | Genera, M, Colcombet-Cazenave, B, Croitoru, A, Raynal, B, Mechaly, A, Caillet, J, Haouz, A, Wolff, N, Caillet-Saguy, C. | | 登録日 | 2023-03-11 | | 公開日 | 2023-05-10 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Interactions of the protein tyrosine phosphatase PTPN3 with viral and cellular partners through its PDZ domain: insights into structural determinants and phosphatase activity.
Front Mol Biosci, 10, 2023
|
|
8KG4
 
 | | Crystal Structure of M- and C-Domains of the shaft pilin LrpA from Ligilactobacillus ruminis - orthorhombic form | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, IODIDE ION, ... | | 著者 | Prajapati, A, Palva, A, von Ossowski, I, Krishnan, V. | | 登録日 | 2023-08-17 | | 公開日 | 2024-07-10 | | 最終更新日 | 2024-07-17 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | The crystal structure of the N-terminal domain of the backbone pilin LrpA reveals a new closure-and-twist motion for assembling dynamic pili in Ligilactobacillus ruminis.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8P3D
 
 | |
8OV8
 
 | | Crystal structure of Ene-reductase 1 from black poplar mushroom in complex to NADP | | 分子名称: | Ene-reductase 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SODIUM ION, ... | | 著者 | Korf, L, Essen, L.-O, Karrer, D, Ruehl, M. | | 登録日 | 2023-04-25 | | 公開日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Shifting the substrate scope of an ene/yne-reductase by loop engineering
To Be Published
|
|
8PE9
 
 | | Complex between DDR1 DS-like domain and PRTH-101 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Liu, J, Chiang, H, Xiong, W, Laurent, V, Griffiths, S.C, Duelfer, J, Deng, H, Sun, X, Yin, Y.W, Li, W, Audoly, L.P, An, Z, Schuerpf, T, Li, R, Zhang, N. | | 登録日 | 2023-06-13 | | 公開日 | 2023-06-28 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (3.152 Å) | | 主引用文献 | A highly selective humanized DDR1 mAb reverses immune exclusion by disrupting collagen fiber alignment in breast cancer.
J Immunother Cancer, 11, 2023
|
|
8OFD
 
 | |
8OJU
 
 | | Crystal structure of the human IgD Fab - structure Fab3 | | 分子名称: | 1,2-ETHANEDIOL, Human IgD Fab heavy chain, Human IgD Fab light chain, ... | | 著者 | Davies, A.M, Beavil, R.L, McDonnell, J.M. | | 登録日 | 2023-03-24 | | 公開日 | 2023-06-14 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Crystal structures of the human IgD Fab reveal insights into C H 1 domain diversity.
Mol.Immunol., 159, 2023
|
|
8OJT
 
 | | Crystal structure of the human IgD Fab - structure Fab2 | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AZIDE ION, ... | | 著者 | Davies, A.M, Beavil, R.L, McDonnell, J.M. | | 登録日 | 2023-03-24 | | 公開日 | 2023-06-14 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Crystal structures of the human IgD Fab reveal insights into C H 1 domain diversity.
Mol.Immunol., 159, 2023
|
|
8I4D
 
 | | X-ray structure of a L-rhamnose-alpha-1,4-D-glucuronate lyase from Fusarium oxysporum 12S, L-Rha complex at 100K | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CALCIUM ION, ... | | 著者 | Yano, N, Kondo, T, Kusaka, K, Yamada, T, Arakawa, T, Sakamoto, T, Fushinobu, S. | | 登録日 | 2023-01-19 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.06 Å) | | 主引用文献 | Charge neutralization and beta-elimination cleavage mechanism of family 42 L-rhamnose-alpha-1,4-D-glucuronate lyase revealed using neutron crystallography.
J.Biol.Chem., 300, 2024
|
|
8PHA
 
 | | O(S)-methyltransferase from Pleurotus sapidus | | 分子名称: | 2-HYDROXY BUTANE-1,4-DIOL, GLYCEROL, L-ornithine, ... | | 著者 | Korf, L, Essen, L.-O. | | 登録日 | 2023-06-19 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | A Novel O - and S -Methyltransferase from Pleurotus sapidus Is Involved in Flavor Formation.
J.Agric.Food Chem., 72, 2024
|
|
8HY5
 
 | | Structure of D-amino acid oxidase mutant R38H | | 分子名称: | 1,2-ETHANEDIOL, BENZOIC ACID, D-amino-acid oxidase, ... | | 著者 | Khan, S, Upadhyay, S, Dave, U, Kumar, A, Gomes, J. | | 登録日 | 2023-01-05 | | 公開日 | 2023-01-25 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural and mechanistic insights into ALS patient derived mutations in D-amino acid oxidase.
Int.J.Biol.Macromol., 256, 2023
|
|
1NAH
 
 | | UDP-GALACTOSE 4-EPIMERASE FROM ESCHERICHIA COLI, REDUCED | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Thoden, J.B, Frey, P.A, Holden, H.M. | | 登録日 | 1995-11-22 | | 公開日 | 1996-12-23 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structures of the oxidized and reduced forms of UDP-galactose 4-epimerase isolated from Escherichia coli.
Biochemistry, 35, 1996
|
|
8P5A
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 obtained in presence of 5 millimolar X77 enantiomer R. | | 分子名称: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, CHLORIDE ION, ... | | 著者 | Costanzi, E, Demitri, N, Storici, P. | | 登録日 | 2023-05-23 | | 公開日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
8P55
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 obtained in presence of 75 micromolar MG-132. | | 分子名称: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, CHLORIDE ION, ... | | 著者 | Costanzi, E, Demitri, N, Storici, P. | | 登録日 | 2023-05-23 | | 公開日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
8P57
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 obtained in presence of 75 micromolar X77. | | 分子名称: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, CHLORIDE ION, ... | | 著者 | Costanzi, E, Demitri, N, Storici, P. | | 登録日 | 2023-05-23 | | 公開日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
8P5C
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 obtained in presence of 5 millimolar X77 enantiomer S. | | 分子名称: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ACETATE ION, ... | | 著者 | Costanzi, E, Demitri, N, Storici, P. | | 登録日 | 2023-05-23 | | 公開日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
8P5B
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 obtained in presence of 500 micromolar X77 enantiomer S. | | 分子名称: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, CHLORIDE ION, ... | | 著者 | Costanzi, E, Demitri, N, Storici, P. | | 登録日 | 2023-05-23 | | 公開日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
8P56
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 obtained in presence of 150 micromolar X77. | | 分子名称: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, CHLORIDE ION, ... | | 著者 | Costanzi, E, Demitri, N, Storici, P. | | 登録日 | 2023-05-23 | | 公開日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
