2HMO
 
 | | Crystal Structure of Naphthalene 1,2-Dioxygenase Bound to 3-Nitrotoluene. | | 分子名称: | 1,2-ETHANEDIOL, 3-NITROTOLUENE, FE (III) ION, ... | | 著者 | Ferraro, D.J, Okerlund, A.L, Mowers, J.C, Ramaswamy, S. | | 登録日 | 2006-07-11 | | 公開日 | 2006-10-10 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural basis for regioselectivity and stereoselectivity of product formation by naphthalene 1,2-dioxygenase.
J.Bacteriol., 188, 2006
|
|
2HMK
 
 | | Crystal Structure of Naphthalene 1,2-Dioxygenase Bound to Phenanthrene | | 分子名称: | 1,2-ETHANEDIOL, FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | 著者 | Ferraro, D.J, Okerlund, A.L, Mowers, J.C, Ramaswamy, S. | | 登録日 | 2006-07-11 | | 公開日 | 2006-10-10 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural basis for regioselectivity and stereoselectivity of product formation by naphthalene 1,2-dioxygenase.
J.Bacteriol., 188, 2006
|
|
3TJH
 
 | | 42F3-p3A1/H2-Ld complex | | 分子名称: | 42F3 alpha, 42F3 beta, H2-Ld SBM2, ... | | 著者 | Adams, J.J, Kruse, A, Kranz, D.M, Garcia, K.C. | | 登録日 | 2011-08-24 | | 公開日 | 2011-12-07 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | T cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex.
Immunity, 35, 2011
|
|
2DD1
 
 | | Three consecutive sheared GA pairs in 5'GGUGGAGGCU/3'PCCAAAGCCG | | 分子名称: | 5'-R(*GP*CP*CP*GP*AP*AP*AP*CP*CP*(P5P))-3', 5'-R(*GP*GP*UP*GP*GP*AP*GP*GP*CP*U)-3' | | 著者 | Chen, G, Kennedy, S.D, Krugh, T.R, Turner, D.H. | | 登録日 | 2006-01-19 | | 公開日 | 2006-06-13 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | An Alternating Sheared AA Pair and Elements of Stability for a Single Sheared Purine-Purine Pair Flanked by Sheared GA Pairs in RNA
Biochemistry, 45, 2006
|
|
3PBB
 
 | | Crystal structure of human secretory glutaminyl cyclase in complex with PBD150 | | 分子名称: | 1-(3,4-dimethoxyphenyl)-3-[3-(1H-imidazol-1-yl)propyl]thiourea, Glutaminyl-peptide cyclotransferase, ZINC ION | | 著者 | Huang, K.F, Liaw, S.S, Huang, W.L, Chia, C.Y, Lo, Y.C, Chen, Y.L, Wang, A.H.J. | | 登録日 | 2010-10-20 | | 公開日 | 2011-02-02 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structures of human Golgi-resident glutaminyl cyclase and its complexes with inhibitors reveal a large loop movement upon inhibitor binding
J.Biol.Chem., 286, 2011
|
|
4I7N
 
 | |
5DLY
 
 | |
7ZXU
 
 | | SARS-CoV-2 Omicron BA.4/5 RBD in complex with Beta-27 Fab and C1 nanobody | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-27 heavy chain, Beta-27 light chain, ... | | 著者 | Huo, J, Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2022-05-23 | | 公開日 | 2022-06-29 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Antibody escape of SARS-CoV-2 Omicron BA.4 and BA.5 from vaccine and BA.1 serum.
Cell, 185, 2022
|
|
2R55
 
 | | Human StAR-related lipid transfer protein 5 | | 分子名称: | StAR-related lipid transfer protein 5 | | 著者 | Lehtio, L, Busam, R.D, Arrowsmith, C.H, Berglund, H, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Herman, M.D, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Moche, M, Nordlund, P, Nyman, T, Sagemark, J, Sundstrom, M, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Persson, C, Structural Genomics Consortium (SGC) | | 登録日 | 2007-09-03 | | 公開日 | 2007-09-18 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Comparative structural analysis of lipid binding START domains.
Plos One, 6, 2011
|
|
3P0C
 
 | | Nischarin PX-domain | | 分子名称: | GLYCEROL, Nischarin | | 著者 | Schutz, P, Karlberg, T, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kol, S, Kotenyova, T, Kouznetsova, E, Moche, M, Nyman, T, Persson, C, Siponen, M.I, Thorsell, A.G, Tresaugues, L, Van Der Berg, S, Wahlberg, E, Welin, M, Weigelt, J, Nordlund, P, Schuler, H, Structural Genomics Consortium (SGC) | | 登録日 | 2010-09-28 | | 公開日 | 2010-10-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.273 Å) | | 主引用文献 | Crystal structure of Nischarin PX-domain
to be published
|
|
2HMN
 
 | | Crystal Structure of the Naphthalene 1,2-Dioxygenase F352V Mutant Bound to Anthracene. | | 分子名称: | 1,2-ETHANEDIOL, ANTHRACENE, FE (III) ION, ... | | 著者 | Ferraro, D.J, Okerlund, A.L, Mowers, J.C, Ramaswamy, S. | | 登録日 | 2006-07-11 | | 公開日 | 2006-10-10 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural basis for regioselectivity and stereoselectivity of product formation by naphthalene 1,2-dioxygenase.
J.Bacteriol., 188, 2006
|
|
8A4Y
 
 | | SARS-CoV-2 non-structural protein-1 (nsp1) in complex with N-(2,3-dihydro-1H-inden-5-yl)acetamide | | 分子名称: | Host translation inhibitor nsp1, N-(2,3-dihydro-1H-inden-5-yl)acetamide | | 著者 | Borsatto, A, Galdadas, I, Ma, S, Damfo, S, Haider, S, Kozielski, F, Estarellas, C, Gervasio, F.L. | | 登録日 | 2022-06-13 | | 公開日 | 2022-11-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.099 Å) | | 主引用文献 | Revealing druggable cryptic pockets in the Nsp1 of SARS-CoV-2 and other beta-coronaviruses by simulations and crystallography.
Elife, 11, 2022
|
|
3P3R
 
 | |
8A55
 
 | |
4I7O
 
 | |
4I7M
 
 | |
2I17
 
 | | Human aldose reductase in complex with NADP+ and the inhibitor IDD594 at temperature of 60K | | 分子名称: | Aldose reductase, CITRIC ACID, IDD594, ... | | 著者 | Petrova, T, Ginell, S, Mitshler, A, Hasemann, I, Schneider, T, Cousido, A, Lunin, V.Y, Joachimiak, A, Podjarny, A. | | 登録日 | 2006-08-13 | | 公開日 | 2006-08-29 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (0.81 Å) | | 主引用文献 | Ultrahigh-resolution study of protein atomic displacement parameters at cryotemperatures obtained with a helium cryostat.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
5ECE
 
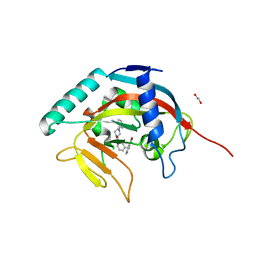 | | Tankyrase 1 with Phthalazinone 1 | | 分子名称: | 1,2-ETHANEDIOL, 2-[4-[3-[(4-oxidanylidene-3~{H}-phthalazin-1-yl)methyl]phenyl]carbonylpiperazin-1-yl]pyridine-3-carbonitrile, ACETATE ION, ... | | 著者 | Kazmirski, S.L, Johannes, J, Read, J.A, Howard, T, Larsen, N.A, Ferguson, A.D. | | 登録日 | 2015-10-20 | | 公開日 | 2015-11-25 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Discovery of AZ0108, an orally bioavailable phthalazinone PARP inhibitor that blocks centrosome clustering.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
5DZX
 
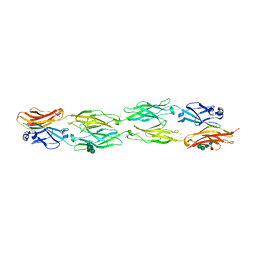 | | Protocadherin beta 6 extracellular cadherin domains 1-4 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Protocadherin beta 6, ... | | 著者 | Goodman, K.M, Mannepalli, S, Bahna, F, Honig, B, Shapiro, L. | | 登録日 | 2015-09-26 | | 公開日 | 2016-05-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.879 Å) | | 主引用文献 | Structural Basis of Diverse Homophilic Recognition by Clustered alpha- and beta-Protocadherins.
Neuron, 90, 2016
|
|
2BV1
 
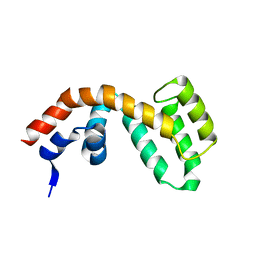 | | Regulator of G-protein Signalling 1 (Human) | | 分子名称: | REGULATOR OF G-PROTEIN SIGNALLING 1 | | 著者 | Elkins, J.M, Yang, X, Soundararajan, M, Schoch, G.A, Haroniti, A, Sundstrom, M, Edwards, A, Arrowsmith, C, Doyle, D.A. | | 登録日 | 2005-06-20 | | 公開日 | 2005-06-27 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha-subunits.
Proc. Natl. Acad. Sci. U.S.A., 105, 2008
|
|
2I16
 
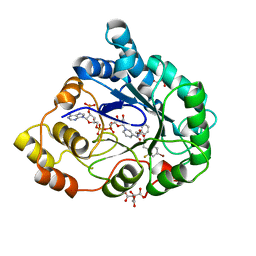 | | Human aldose reductase in complex with NADP+ and the inhibitor IDD594 at temperature of 15K | | 分子名称: | Aldose reductase, CITRIC ACID, IDD594, ... | | 著者 | Petrova, T, Ginell, S, Mitshler, A, Hasemann, I, Schneider, T, Cousido, A, Lunin, V.Y, Joachimiak, A, Podjarny, A. | | 登録日 | 2006-08-13 | | 公開日 | 2006-08-29 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (0.81 Å) | | 主引用文献 | Ultrahigh-resolution study of protein atomic displacement parameters at cryotemperatures obtained with a helium cryostat.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2BY9
 
 | | Is radiation damage dependent on the dose-rate used during macromolecular crystallography data collection | | 分子名称: | BENZAMIDINE, CALCIUM ION, CATIONIC TRYPSIN, ... | | 著者 | Leiros, H.-K.S, Timmins, J, Ravelli, R.B.G, McSweeney, S.M. | | 登録日 | 2005-07-29 | | 公開日 | 2006-02-06 | | 最終更新日 | 2021-08-04 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Is Radiation Damage Dependent on the Dose-Rate Used During Macromolecular Crystallography Data Collection?
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2BY8
 
 | | Is radiation damage dependent on the dose-rate used during macromolecular crystallography data collection | | 分子名称: | BENZAMIDINE, CALCIUM ION, CATIONIC TRYPSIN, ... | | 著者 | Leiros, H.-K.S, Timmins, J, Ravelli, R.B.G, McSweeney, S.M. | | 登録日 | 2005-07-29 | | 公開日 | 2006-02-06 | | 最終更新日 | 2021-08-04 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Is Radiation Damage Dependent on the Dose-Rate Used During Macromolecular Crystallography Data Collection?
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2BY5
 
 | | Is radiation damage dependent on the dose-rate used during macromolecular crystallography data collection | | 分子名称: | BENZAMIDINE, CALCIUM ION, CATIONIC TRYPSIN, ... | | 著者 | Leiros, H.-K.S, Timmins, J, Ravelli, R.B.G, McSweeney, S.M. | | 登録日 | 2005-07-29 | | 公開日 | 2006-02-06 | | 最終更新日 | 2021-08-04 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Is Radiation Damage Dependent on the Dose-Rate Used During Macromolecular Crystallography Data Collection?
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2C5K
 
 | | N-terminal domain of tlg1 complexed with N-terminus of vps51 | | 分子名称: | SULFATE ION, T-SNARE AFFECTING A LATE GOLGI COMPARTMENT PROTEIN 1, VACUOLAR PROTEIN SORTING PROTEIN 51 | | 著者 | Fridmann-Sirkis, Y, Kent, H.M, Lewis, M.J, Evans, P.R, Pelham, H.R.B. | | 登録日 | 2005-10-27 | | 公開日 | 2006-01-25 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structural Analysis of the Interaction between the Snare Tlg1 and Vps51.
Traffic, 7, 2006
|
|
