4BRG
 
 | | Legionella pneumophila NTPDase1 crystal form II (closed) in complex with MG GMPPNP | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE I, ... | | 著者 | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | 登録日 | 2013-06-04 | | 公開日 | 2013-07-17 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Crystallographic Snapshots Along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases
Structure, 21, 2013
|
|
4BRQ
 
 | | LEGIONELLA PNEUMOPHILA NTPDASE1 CRYSTAL FORM II, CLOSED, IN COMPLEX WITH TWO PHOSPHATES BOUND TO ACTIVE SITE MG AND PRODUCT AMP | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | 著者 | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | 登録日 | 2013-06-05 | | 公開日 | 2013-07-17 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Crystallographic Snapshots Along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases
Structure, 21, 2013
|
|
7XFH
 
 | | Structure of nucleosome-AAG complex (A-30I, post-catalytic state) | | 分子名称: | DNA (152-MER), DNA-3-methyladenine glycosylase, Histone H2A type 1, ... | | 著者 | Zheng, L, Tsai, B, Gao, N. | | 登録日 | 2022-04-01 | | 公開日 | 2023-04-19 | | 最終更新日 | 2025-06-25 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural and mechanistic insights into the DNA glycosylase AAG-mediated base excision in nucleosome.
Cell Discov, 9, 2023
|
|
3HVT
 
 | | STRUCTURAL BASIS OF ASYMMETRY IN THE HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 REVERSE TRANSCRIPTASE HETERODIMER | | 分子名称: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P51), HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P66) | | 著者 | Steitz, T.A, Smerdon, S.J, Jaeger, J, Wang, J, Kohlstaedt, L.A, Chirino, A.J, Friedman, J.M, Rice, P.A. | | 登録日 | 1994-07-25 | | 公開日 | 1994-10-15 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structure of the binding site for nonnucleoside inhibitors of the reverse transcriptase of human immunodeficiency virus type 1.
Proc.Natl.Acad.Sci.Usa, 91, 1994
|
|
7XNP
 
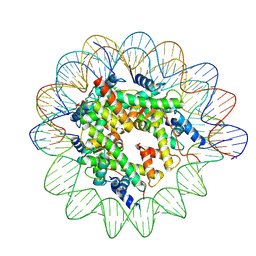 | | Structure of nucleosome-AAG complex (A-55I, post-catalytic state) | | 分子名称: | DNA (152-MER), Histone H2A type 1, Histone H2B 1.1, ... | | 著者 | Zheng, L, Tsai, B, Gao, N. | | 登録日 | 2022-04-29 | | 公開日 | 2023-05-03 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural and mechanistic insights into the DNA glycosylase AAG-mediated base excision in nucleosome.
Cell Discov, 9, 2023
|
|
6LA8
 
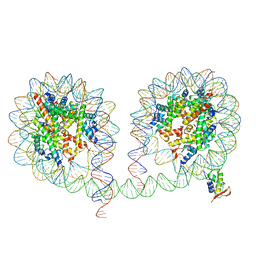 | | 349 bp di-nucleosome harboring cohesive DNA termini assembled with linker histone H1.0 | | 分子名称: | CALCIUM ION, DNA (349-MER), Histone H1.0, ... | | 著者 | Adhireksan, Z, Lee, P.L, Sharma, D, Davey, C.A. | | 登録日 | 2019-11-12 | | 公開日 | 2020-10-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Near-atomic resolution structures of interdigitated nucleosome fibres.
Nat Commun, 11, 2020
|
|
7VDV
 
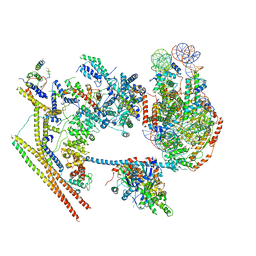 | | The overall structure of human chromatin remodeling PBAF-nucleosome complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, AT-rich interactive domain-containing protein 2,AT-rich interactive domain-containing protein 2, Actin, ... | | 著者 | Chen, Z.C, Chen, K.J, Yuan, J.J. | | 登録日 | 2021-09-07 | | 公開日 | 2022-05-18 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure of human chromatin-remodelling PBAF complex bound to a nucleosome.
Nature, 605, 2022
|
|
6ZHX
 
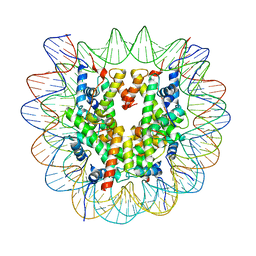 | | Cryo-EM structure of the regulatory linker of ALC1 bound to the nucleosome's acidic patch: nucleosome class. | | 分子名称: | Chromodomain-helicase-DNA-binding protein 1-like, DNA (145-MER) Widom 601 sequence, Histone H2A type 1, ... | | 著者 | Bacic, L, Gaullier, G, Croll, T.I, Deindl, S. | | 登録日 | 2020-06-24 | | 公開日 | 2020-12-23 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Mechanistic Insights into Regulation of the ALC1 Remodeler by the Nucleosome Acidic Patch.
Cell Rep, 33, 2020
|
|
6LA9
 
 | | 349 bp di-nucleosome harboring cohesive DNA termini assembled with linker histone H1.0 (high cryoprotectant) | | 分子名称: | CALCIUM ION, DNA (349-MER), Histone H1.0, ... | | 著者 | Adhireksan, Z, Sharma, D, Lee, P.L, Davey, C.A. | | 登録日 | 2019-11-12 | | 公開日 | 2020-10-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Near-atomic resolution structures of interdigitated nucleosome fibres.
Nat Commun, 11, 2020
|
|
6KW4
 
 | | The ClassB RSC-Nucleosome Complex | | 分子名称: | Actin-like protein ARP9, Actin-related protein 7, Chromatin structure-remodeling complex protein RSC3, ... | | 著者 | Ye, Y.P, Wu, H, Chen, K.J, Verma, N, Cairns, B, Gao, N, Chen, Z.C. | | 登録日 | 2019-09-06 | | 公開日 | 2019-11-13 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (7.55 Å) | | 主引用文献 | Structure of the RSC complex bound to the nucleosome.
Science, 366, 2019
|
|
8JND
 
 | | The cryo-EM structure of the nonameric RAD51 ring bound to the nucleosome with the linker DNA binding | | 分子名称: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-06-06 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.66 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
7WBV
 
 | | RNA polymerase II elongation complex bound with Elf1 and Spt4/5, stalled at SHL(-4) of the nucleosome | | 分子名称: | DNA (159-MER), DNA (198-MER), DNA-directed RNA polymerase subunit, ... | | 著者 | Osumi, K, Kujirai, T, Ehara, H, Sekine, S, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2021-12-17 | | 公開日 | 2023-07-05 | | 最終更新日 | 2025-06-25 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structural Basis of Damaged Nucleotide Recognition by Transcribing RNA Polymerase II in the Nucleosome.
J.Mol.Biol., 435, 2023
|
|
7UND
 
 | | Pol II-DSIF-SPT6-PAF1c-TFIIS-nucleosome complex (stalled at +38) | | 分子名称: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB3, DNA-directed RNA polymerase II subunit RPB7, ... | | 著者 | Filipovski, M, Vos, S.M, Farnung, L. | | 登録日 | 2022-04-10 | | 公開日 | 2022-10-19 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis of nucleosome retention during transcription elongation.
Science, 376, 2022
|
|
8W9F
 
 | |
8W9E
 
 | |
8W9D
 
 | |
7XFL
 
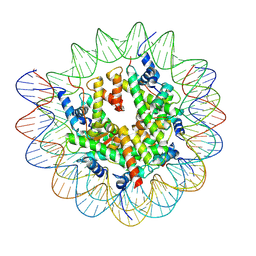 | | Structure of nucleosome-AAG complex (A-53I, free state) | | 分子名称: | DNA (152-MER), Histone H2A type 1, Histone H2B 1.1, ... | | 著者 | Zheng, L, Tsai, B, Gao, N. | | 登録日 | 2022-04-01 | | 公開日 | 2023-04-19 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structural and mechanistic insights into the DNA glycosylase AAG-mediated base excision in nucleosome.
Cell Discov, 9, 2023
|
|
4G89
 
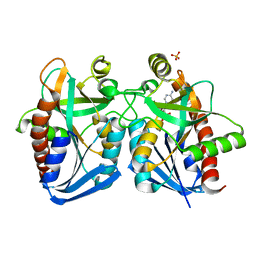 | | Crystal structure of k. pneumoniae mta/adohcy nucleosidase in complex with fragmented s-adenosyl-l-homocysteine | | 分子名称: | 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, ADENINE, S-ADENOSYL-L-HOMOCYSTEINE, ... | | 著者 | Norris, G.E, Brown, R.L, Anderson, B.F, Tyler, P.C, Evans, G.B. | | 登録日 | 2012-07-23 | | 公開日 | 2012-09-19 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of k. pneumoniae mta/adohcy nucleosidase in complex with fragmented s-adenosyl-l-homocysteine
To be Published
|
|
6E1C
 
 | |
8XBW
 
 | | The cryo-EM structure of the RAD51 N-terminal lobe domain bound to the histone H4 tail of the nucleosome | | 分子名称: | DNA (5'-D(P*AP*CP*CP*GP*CP*TP*TP*AP*AP*AP*CP*GP*CP*AP*CP*GP*TP*A)-3'), DNA (5'-D(P*TP*AP*CP*GP*TP*GP*CP*GP*TP*TP*TP*AP*AP*GP*CP*GP*GP*T)-3'), DNA repair protein RAD51 homolog 1, ... | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-12-07 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (2.89 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8JNE
 
 | | The cryo-EM structure of the decameric RAD51 ring bound to the nucleosome without the linker DNA binding | | 分子名称: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-06-06 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.68 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8JNF
 
 | | The cryo-EM structure of the RAD51 filament bound to the nucleosome | | 分子名称: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-06-06 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (6.91 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
4BR0
 
 | | rat NTPDase2 in complex with Ca AMPNP | | 分子名称: | 5'-O-[(R)-hydroxy(phosphonoamino)phosphoryl]adenosine, CALCIUM ION, ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE 2, ... | | 著者 | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | 登録日 | 2013-06-03 | | 公開日 | 2013-07-17 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Crystallographic Snapshots Along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases
Structure, 21, 2013
|
|
3MMS
 
 | | Crystal structure of Streptococcus pneumoniae MTA/SAH nucleosidase in complex with 8-aminoadenine | | 分子名称: | 5'-methylthioadenosine / S-adenosylhomocysteine nucleosidase, 9H-purine-6,8-diamine, GLYCEROL | | 著者 | Siu, K.K.W, Lee, J.E, Horvatin-Mrakovcic, C, Howell, P.L. | | 登録日 | 2010-04-20 | | 公開日 | 2010-05-12 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structure of Streptococcus pneumoniae MTA/SAH nucleosidase in complex with 8-aminoadenine
TO BE PUBLISHED
|
|
9JAO
 
 | | The structure of SMARCAD1 bound to the hexasome in the presence of ADP-BeFx | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (157-MER), ... | | 著者 | Chen, Z.C, Hu, P.J, Sia, Y.Y, Chen, K.J, Xia, X. | | 登録日 | 2024-08-25 | | 公開日 | 2025-04-30 | | 最終更新日 | 2025-06-18 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Subnucleosome preference of human chromatin remodeller SMARCAD1.
Nature, 2025
|
|
