2C8F
 
 | | Structure of the ARTT motif E214N mutant C3bot1 Exoenzyme (NAD-bound state, crystal form III) | | 分子名称: | MONO-ADP-RIBOSYLTRANSFERASE C3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Stura, E.A, Menetrey, J, Flatau, G, Boquet, P, Menez, A. | | 登録日 | 2005-12-03 | | 公開日 | 2007-02-27 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural Basis for the Nad-Hydrolysis Mechanism and the Artt-Loop Plasticity of C3 Exoenzymes.
Protein Sci., 17, 2008
|
|
2C8A
 
 | | Structure of the wild-type C3bot1 Exoenzyme (Nicotinamide-bound state, crystal form I) | | 分子名称: | MONO-ADP-RIBOSYLTRANSFERASE C3, NICOTINAMIDE, SULFATE ION | | 著者 | Stura, E.A, Menetrey, J, Flatau, G, Boquet, P, Menez, A. | | 登録日 | 2005-12-03 | | 公開日 | 2007-02-27 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural Basis for the Nad-Hydrolysis Mechanism and the Artt-Loop Plasticity of C3 Exoenzymes.
Protein Sci., 17, 2008
|
|
2C8C
 
 | | Structure of the ARTT motif Q212A mutant C3bot1 Exoenzyme (NAD-bound state, crystal form I) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MONO-ADP-RIBOSYLTRANSFERASE C3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Stura, E.A, Menetrey, J, Flatau, G, Boquet, P, Menez, A. | | 登録日 | 2005-12-03 | | 公開日 | 2007-02-27 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural Basis for the Nad-Hydrolysis Mechanism and the Artt-Loop Plasticity of C3 Exoenzymes.
Protein Sci., 17, 2008
|
|
2CIA
 
 | | human nck2 sh2-domain in complex with a decaphosphopeptide from translocated intimin receptor (tir) of epec | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, CYTOPLASMIC PROTEIN NCK2, ISOPROPYL ALCOHOL, ... | | 著者 | Frese, S, Schubert, W.-D, Findeis, A.C, Marquardt, T, Roske, Y.S, Stradal, T.E.B, Heinz, D.W. | | 登録日 | 2006-03-17 | | 公開日 | 2006-04-24 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | The Phosphotyrosine Peptide Binding Specificity of Nck1 and Nck2 Src Homology 2 Domains.
J.Biol.Chem., 281, 2006
|
|
6ZON
 
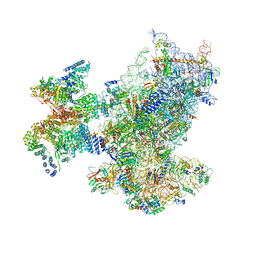 | | SARS-CoV-2 Nsp1 bound to a human 43S preinitiation ribosome complex - state 1 | | 分子名称: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | 著者 | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | 登録日 | 2020-07-07 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
6ZP4
 
 | | SARS-CoV-2 Nsp1 bound to a human 43S preinitiation ribosome complex - state 2 | | 分子名称: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | 著者 | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | 登録日 | 2020-07-08 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
6ZMW
 
 | | Structure of a human 48S translational initiation complex | | 分子名称: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | 著者 | Brito Querido, J, Sokabe, M, Kraatz, S, Gordiyenko, Y, Skehel, M, Fraser, C, Ramakrishnan, V. | | 登録日 | 2020-07-04 | | 公開日 | 2020-09-23 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structure of a human 48Stranslational initiation complex.
Science, 369, 2020
|
|
1OSV
 
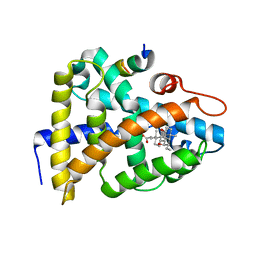 | | STRUCTURAL BASIS FOR BILE ACID BINDING AND ACTIVATION OF THE NUCLEAR RECEPTOR FXR | | 分子名称: | 6-ETHYL-CHENODEOXYCHOLIC ACID, Bile acid receptor, Nuclear receptor coactivator 2 | | 著者 | Mi, L.Z, Devarakonda, S, Harp, J.M, Han, Q, Pellicciari, R, Willson, T.M, Khorasanizadeh, S, Rastinejad, F. | | 登録日 | 2003-03-20 | | 公開日 | 2004-03-23 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural Basis for Bile Acid Binding and Activation of the Nuclear Receptor FXR
Mol.Cell, 11, 2003
|
|
2DGN
 
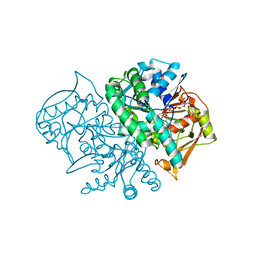 | | Mouse Muscle Adenylosuccinate Synthetase partially ligated complex with GTP, 2'-deoxy-IMP | | 分子名称: | 9-(2-DEOXY-5-O-PHOSPHONO-BETA-D-ERYTHRO-PENTOFURANOSYL)-6-(PHOSPHONOOXY)-9H-PURINE, Adenylosuccinate synthetase isozyme 1, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Iancu, C.V, Zhou, Y, Borza, T, Fromm, H.J, Honzatko, R.B. | | 登録日 | 2006-03-15 | | 公開日 | 2006-09-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Cavitation as a mechanism of substrate discrimination by adenylosuccinate synthetases.
Biochemistry, 45, 2006
|
|
2CLS
 
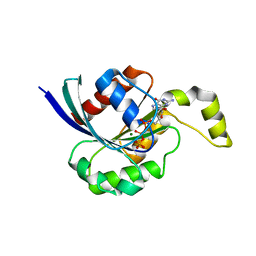 | | The crystal structure of the human RND1 GTPase in the active GTP bound state | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RHO-RELATED GTP-BINDING PROTEIN RHO6 | | 著者 | Pike, A.C.W, Yang, X, Colebrook, S, Gileadi, O, Sobott, F, Bray, J, Wen Hwa, L, Marsden, B, Zhao, Y, Schoch, G, Elkins, J, Debreczeni, J.E, Turnbull, A.P, von Delft, F, Arrowsmith, C, Edwards, A, Weigelt, J, Sundstrom, M, Doyle, D. | | 登録日 | 2006-04-28 | | 公開日 | 2006-05-04 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | The Crystal Structure of the Human Rnd1 Gtpase in the Active GTP Bound State
To be Published
|
|
2C8D
 
 | | Structure of the ARTT motif Q212A mutant C3bot1 Exoenzyme (Free state, crystal form I) | | 分子名称: | MONO-ADP-RIBOSYLTRANSFERASE C3, SULFATE ION | | 著者 | Stura, E.A, Menetrey, J, Flatau, G, Boquet, P, Menez, A. | | 登録日 | 2005-12-03 | | 公開日 | 2007-02-27 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural Basis for the Nad-Hydrolysis Mechanism and the Artt-Loop Plasticity of C3 Exoenzymes.
Protein Sci., 17, 2008
|
|
2C8E
 
 | | Structure of the ARTT motif E214N mutant C3bot1 Exoenzyme (Free state, crystal form III) | | 分子名称: | MONO-ADP-RIBOSYLTRANSFERASE C3, SULFATE ION | | 著者 | Stura, E.A, Menetrey, J, Flatau, G, Boquet, P, Menez, A. | | 登録日 | 2005-12-03 | | 公開日 | 2007-02-27 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural Basis for the Nad-Hydrolysis Mechanism and the Artt-Loop Plasticity of C3 Exoenzymes.
Protein Sci., 17, 2008
|
|
2C8B
 
 | | Structure of the ARTT motif Q212A mutant C3bot1 Exoenzyme (Free state, crystal form II) | | 分子名称: | MONO-ADP-RIBOSYLTRANSFERASE C3, SULFATE ION | | 著者 | Stura, E.A, Menetrey, J, Flatau, G, Boquet, P, Menez, A. | | 登録日 | 2005-12-03 | | 公開日 | 2007-02-27 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural Basis for the Nad-Hydrolysis Mechanism and the Artt-Loop Plasticity of C3 Exoenzymes.
Protein Sci., 17, 2008
|
|
2DLM
 
 | | Solution structure of the first SH3 domain of human vinexin | | 分子名称: | Vinexin | | 著者 | Zhang, H.P, Izumi, K, Yoshida, M, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2006-04-20 | | 公開日 | 2006-10-20 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the first SH3 domain of human vinexin
To be published
|
|
2BBM
 
 | | SOLUTION STRUCTURE OF A CALMODULIN-TARGET PEPTIDE COMPLEX BY MULTIDIMENSIONAL NMR | | 分子名称: | CALCIUM ION, CALMODULIN, MYOSIN LIGHT CHAIN KINASE | | 著者 | Clore, G.M, Bax, A, Ikura, M, Gronenborn, A.M. | | 登録日 | 1992-07-16 | | 公開日 | 1994-01-31 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a calmodulin-target peptide complex by multidimensional NMR.
Science, 256, 1992
|
|
2BBN
 
 | | SOLUTION STRUCTURE OF A CALMODULIN-TARGET PEPTIDE COMPLEX BY MULTIDIMENSIONAL NMR | | 分子名称: | CALCIUM ION, CALMODULIN, MYOSIN LIGHT CHAIN KINASE | | 著者 | Clore, G.M, Bax, A, Ikura, M, Gronenborn, A.M. | | 登録日 | 1992-07-16 | | 公開日 | 1994-01-31 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a calmodulin-target peptide complex by multidimensional NMR.
Science, 256, 1992
|
|
7DHT
 
 | |
4QBM
 
 | | Crystal structure of human BAZ2A bromodomain in complex with a diacetylated histone 4 peptide (H4K16acK20ac) | | 分子名称: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2A, histone H4 peptide with sequence Gly-Ala-Lys(ac)-Arg-His-Arg-Lys(ac)-Val-Leu | | 著者 | Tallant, C, Nunez-Alonso, G, Picaud, S, Filippakopoulos, P, Krojer, T, Williams, E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2014-05-08 | | 公開日 | 2014-05-21 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Molecular basis of histone tail recognition by human TIP5 PHD finger and bromodomain of the chromatin remodeling complex NoRC.
Structure, 23, 2015
|
|
2EMS
 
 | | Crystal Structure Analysis of the radixin FERM domain complexed with adhesion molecule CD43 | | 分子名称: | Leukosialin, Radixin | | 著者 | Takai, Y, Kitano, K, Terawaki, S, Maesaki, R, Hakoshima, T. | | 登録日 | 2007-03-28 | | 公開日 | 2008-04-01 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural basis of the cytoplasmic tail of adhesion molecule CD43 and its binding to ERM proteins
J.Mol.Biol., 381, 2008
|
|
2E2P
 
 | | Crystal structure of Sulfolobus tokodaii hexokinase in complex with ADP | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, HEXOKINASE, ... | | 著者 | Nishimasu, H, Fushinobu, S, Shoun, H, Wakagi, T. | | 登録日 | 2006-11-15 | | 公開日 | 2007-01-16 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structures of an ATP-dependent hexokinase with broad substrate specificity from the hyperthermophilic archaeon Sulfolobus tokodaii.
J.Biol.Chem., 282, 2007
|
|
2EMT
 
 | | Crystal Structure Analysis of the radixin FERM domain complexed with adhesion molecule PSGL-1 | | 分子名称: | P-selectin glycoprotein ligand 1, Radixin | | 著者 | Takai, Y, Kitano, K, Terawaki, S, Maesaki, R, Hakoshima, T. | | 登録日 | 2007-03-28 | | 公開日 | 2008-03-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural basis of PSGL-1 binding to ERM proteins
Genes Cells, 12, 2007
|
|
2AIN
 
 | |
2AHE
 
 | | Crystal structure of a soluble form of CLIC4. intercellular chloride ion channel | | 分子名称: | Chloride intracellular channel protein 4 | | 著者 | Littler, D.R, Assaad, N.N, Harrop, S.J, Brown, L.J, Pankhurst, G.J, Luciani, P, Aguilar, M.-I, Mazzanti, M, Berryman, M.A, Breit, S.N, Curmi, P.M.G. | | 登録日 | 2005-07-28 | | 公開日 | 2005-08-16 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of the soluble form of the redox-regulated chloride ion channel protein CLIC4.
FEBS J., 272, 2005
|
|
2A5Y
 
 | | Structure of a CED-4/CED-9 complex | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Apoptosis regulator ced-9, MAGNESIUM ION, ... | | 著者 | Yan, N, Liu, Q, Hao, Q, Gu, L, Shi, Y. | | 登録日 | 2005-07-01 | | 公開日 | 2005-10-11 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure of the CED-4-CED-9 complex provides insights into programmed cell death in Caenorhabditis elegans.
Nature, 437, 2005
|
|
2E2Q
 
 | | Crystal structure of Sulfolobus tokodaii hexokinase in complex with xylose, Mg2+, and ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, HEXOKINASE, MAGNESIUM ION, ... | | 著者 | Nishimasu, H, Fushinobu, S, Shoun, H, Wakagi, T. | | 登録日 | 2006-11-15 | | 公開日 | 2007-01-16 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structures of an ATP-dependent hexokinase with broad substrate specificity from the hyperthermophilic archaeon Sulfolobus tokodaii.
J.Biol.Chem., 282, 2007
|
|
