3MRH
 
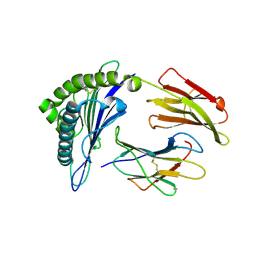 | | Crystal Structure of MHC class I HLA-A2 molecule complexed with HCV NS3-1073-1081 nonapeptide N3S variant | | 分子名称: | 9-meric peptide from Serine protease/NTPase/helicase NS3, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | 著者 | Gras, S, Reiser, J.-B, Chouquet, A, Le Gorrec, M, Debeaupuis, E, Echasserieau, K, Saulquin, X, Bonneville, M, Housset, D. | | 登録日 | 2010-04-29 | | 公開日 | 2011-05-25 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Analysis of Relationships between Peptide/MHC Structural Features and Naive T Cell Frequency in Humans.
J.Immunol., 193, 2014
|
|
6ZD1
 
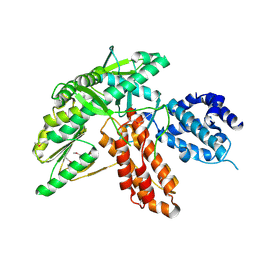 | | Structure of apo telomerase from Candida Tropicalis | | 分子名称: | Telomerase reverse transcriptase | | 著者 | Zhai, L, Rety, S, Chen, W.F, Auguin, D, Xi, X.G. | | 登録日 | 2020-06-13 | | 公開日 | 2021-04-28 | | 最終更新日 | 2021-05-12 | | 実験手法 | X-RAY DIFFRACTION (2.47 Å) | | 主引用文献 | Crystal structures of N-terminally truncated telomerase reverse transcriptase from fungi‡.
Nucleic Acids Res., 49, 2021
|
|
6ZD6
 
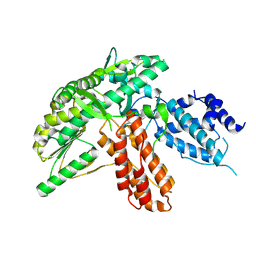 | | Structure of apo telomerase from Candida Tropicalis | | 分子名称: | Telomerase reverse transcriptase | | 著者 | Zhai, L, Rety, S, Chen, W.F, Auguin, D, Xi, X.G. | | 登録日 | 2020-06-13 | | 公開日 | 2021-04-28 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.64 Å) | | 主引用文献 | Crystal structures of N-terminally truncated telomerase reverse transcriptase from fungi‡.
Nucleic Acids Res., 49, 2021
|
|
1A1R
 
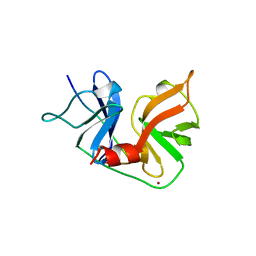 | | HCV NS3 PROTEASE DOMAIN:NS4A PEPTIDE COMPLEX | | 分子名称: | NS3 PROTEIN, NS4A PROTEIN, ZINC ION | | 著者 | Kim, J.L, Morgenstern, K.A, Lin, C, Fox, T, Dwyer, M.D, Landro, J.A, Chambers, S.P, Markland, W, Lepre, C.A, O'Malley, E.T, Harbeson, S.L, Rice, C.M, Murcko, M.A, Caron, P.R, Thomson, J.A. | | 登録日 | 1997-12-15 | | 公開日 | 1998-06-17 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide.
Cell(Cambridge,Mass.), 87, 1996
|
|
7U1A
 
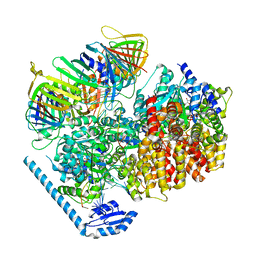 | | RFC:PCNA bound to dsDNA with a ssDNA gap of six nucleotides | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA - Primer, DNA - Template, ... | | 著者 | Liu, X, Gaubitz, C, Pajak, J, Kelch, B.A. | | 登録日 | 2022-02-20 | | 公開日 | 2022-07-06 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | A second DNA binding site on RFC facilitates clamp loading at gapped or nicked DNA.
Elife, 11, 2022
|
|
7U1P
 
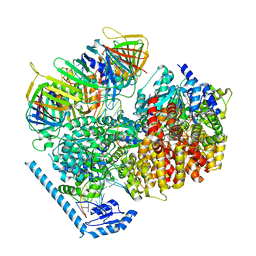 | | RFC:PCNA bound to DNA with a ssDNA gap of five nucleotides | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA - Primer, DNA - Template, ... | | 著者 | Liu, X, Gaubitz, C, Pajak, J, Kelch, B.A. | | 登録日 | 2022-02-21 | | 公開日 | 2022-07-06 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | A second DNA binding site on RFC facilitates clamp loading at gapped or nicked DNA.
Elife, 11, 2022
|
|
1T03
 
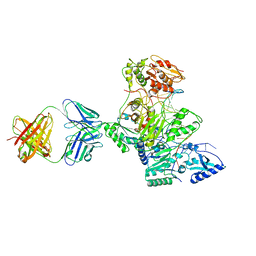 | | HIV-1 reverse transcriptase crosslinked to tenofovir terminated template-primer (complex P) | | 分子名称: | MAGNESIUM ION, POL polyprotein, Synthetic oligonucleotide primer, ... | | 著者 | Tuske, S, Sarafianos, S.G, Ding, J, Arnold, E. | | 登録日 | 2004-04-07 | | 公開日 | 2004-05-11 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structure of HIV-1 RT-DNA complexes before and after incorporation of the anti-AIDS drug tenofovir
Nat.Struct.Mol.Biol., 11, 2004
|
|
1T05
 
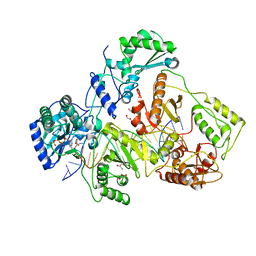 | | HIV-1 reverse transcriptase crosslinked to template-primer with tenofovir-diphosphate bound as the incoming nucleotide substrate | | 分子名称: | GLYCEROL, MAGNESIUM ION, POL polyprotein, ... | | 著者 | Tuske, S, Sarafianos, S.G, Ding, J, Arnold, E. | | 登録日 | 2004-04-07 | | 公開日 | 2004-05-11 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structures of HIV-1 RT-DNA complexes before and after incorporation of the anti-AIDS drug tenofovir
Nat.Struct.Mol.Biol., 11, 2004
|
|
2YMN
 
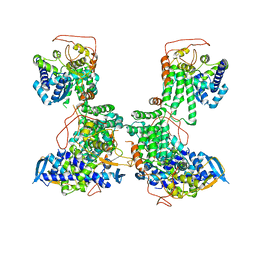 | | Organization of the Influenza Virus Replication Machinery | | 分子名称: | NUCLEOPROTEIN | | 著者 | Moeller, A, Kirchdoerfer, R.N, Potter, C.S, Carragher, B, Wilson, I.A. | | 登録日 | 2012-10-09 | | 公開日 | 2012-12-05 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (20 Å) | | 主引用文献 | Organization of the Influenza Virus Replication Machinery.
Science, 338, 2012
|
|
5T1V
 
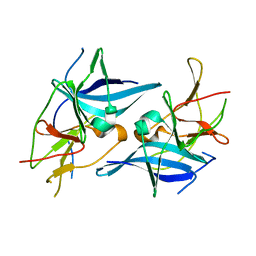 | | Crystal structure of Zika virus NS2B-NS3 protease in apo-form. | | 分子名称: | NS2B-NS3 protease,NS2B-NS3 protease | | 著者 | Nocadello, S, Light, S.H, Minasov, G, Shuvalova, L, Cardona-Correa, A.A, Ojeda, I, Vargas, J, Johnson, M.E, Lee, H, Anderson, W.F, Center for Structural Genomics of Infectious Diseases, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2016-08-22 | | 公開日 | 2016-09-07 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Crystal structure of Zika virus NS2B-NS3 protease in apo-form.
To Be Published
|
|
2CLZ
 
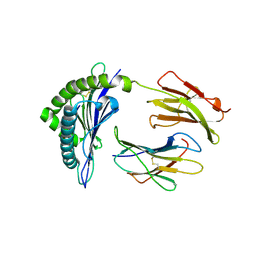 | | Mhc Class I Natural Mutant H-2Kbm8 Heavy Chain Complexed With beta-2 Microglobulin and pBM1 peptide | | 分子名称: | BETA-2 MICROGLOBULIN, H-2 CLASS I HISTOCOMPATIBILITY ANTIGEN, K-B ALPHA CHAIN, ... | | 著者 | Mazza, C, Auphan-Anezin, N, Guimezanes, A, Barrett-Wilt, G.A, Montero-Julian, F, Roussel, A, Hunt, D.F, Schmitt-Verhulst, A.M, Malissen, B. | | 登録日 | 2006-05-03 | | 公開日 | 2006-06-14 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Distinct Orientation of the Alloreactive Monoclonal Cd8 T Cell Activation Program by Three Different Peptide/Mhc Complexes.
Eur.J.Immunol., 36, 2006
|
|
7LUN
 
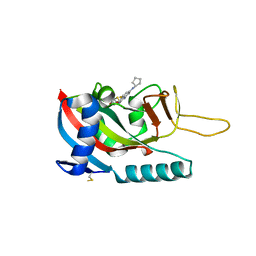 | | Human PARP14 (ARTD8), catalytic fragment in complex with RBN011980 | | 分子名称: | 7-(cyclopentylamino)-5-fluoro-2-{[(trans-4-hydroxycyclohexyl)sulfanyl]methyl}quinazolin-4(3H)-one, CHLORIDE ION, GLYCEROL, ... | | 著者 | Dorsey, B.W, Swinger, K.K, Schenkel, L.B, Church, W.D, Perl, N.R, Vasbinder, M.M, Wigle, T.J, Kuntz, K.W. | | 登録日 | 2021-02-22 | | 公開日 | 2021-04-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.57 Å) | | 主引用文献 | Targeted Degradation of PARP14 Using a Heterobifunctional Small Molecule.
Chembiochem, 22, 2021
|
|
1TL3
 
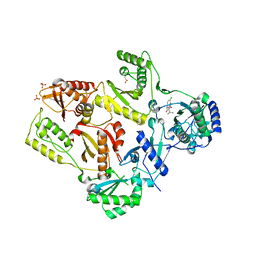 | | Crystal structure of hiv-1 reverse transcriptase in complex with gw450557 | | 分子名称: | 6-CHLORO-4-(CYCLOHEXYLOXY)-3-ISOPROPYLQUINOLIN-2(1H)-ONE, PHOSPHATE ION, Pol polyprotein, ... | | 著者 | Hopkins, A.L, Ren, J, Stuart, D.I, Stammers, D.K. | | 登録日 | 2004-06-09 | | 公開日 | 2004-12-07 | | 最終更新日 | 2022-12-21 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Design of non-nucleoside inhibitors of HIV-1 reverse transcriptase with improved drug resistance properties. 1.
J.Med.Chem., 47, 2004
|
|
1OK8
 
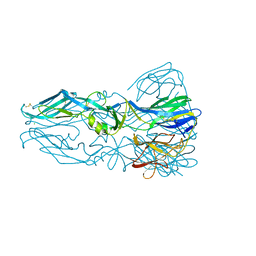 | |
1TKT
 
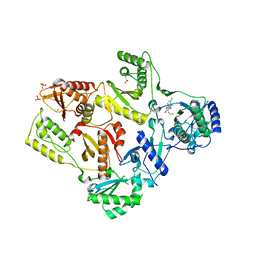 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH GW426318 | | 分子名称: | 6-CHLORO-4-(CYCLOHEXYLOXY)-3-PROPYLQUINOLIN-2(1H)-ONE, MAGNESIUM ION, PHOSPHATE ION, ... | | 著者 | Hopkins, A.L, Ren, J, Stuart, D.I, Stammers, D.K. | | 登録日 | 2004-06-09 | | 公開日 | 2004-12-07 | | 最終更新日 | 2018-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Design of non-nucleoside inhibitors of HIV-1 reverse transcriptase with improved drug resistance properties. 1.
J.Med.Chem., 47, 2004
|
|
1TL1
 
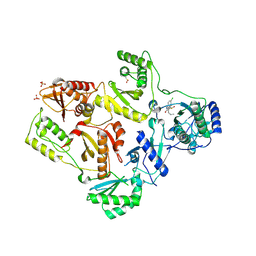 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH GW451211 | | 分子名称: | 6-CHLORO-4-(CYCLOHEXYLSULFINYL)-3-PROPYLQUINOLIN-2(1H)-ONE, PHOSPHATE ION, Pol polyprotein, ... | | 著者 | Hopkins, A.L, Ren, J, Stuart, D.I, Stammers, D.K. | | 登録日 | 2004-06-09 | | 公開日 | 2004-12-07 | | 最終更新日 | 2022-12-21 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Design of non-nucleoside inhibitors of HIV-1 reverse transcriptase with improved drug resistance properties. 1.
J.Med.Chem., 47, 2004
|
|
1DY9
 
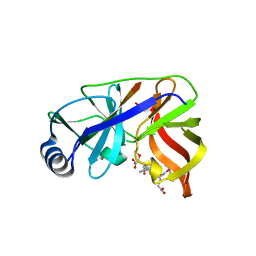 | | Inhibition of the Hepatitis C Virus NS3/4A Protease. The Crystal Structures of Two Protease-Inhibitor Complexes (inhibitor I) | | 分子名称: | N-(tert-butoxycarbonyl)-L-alpha-glutamyl-N-[(1R)-1-(carboxycarbonyl)-3,3-difluoropropyl]-L-leucinamide, NONSTRUCTURAL PROTEIN NS4A (P4), PROTEASE/HELICASE NS3 (P70), ... | | 著者 | Di Marco, S, Rizzi, M, Volpari, C, Walsh, M, Narjes, F, Colarusso, S, De Francesco, R, Matassa, V.G, Sollazzo, M. | | 登録日 | 2000-01-31 | | 公開日 | 2001-01-28 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Inhibition of the Hepatitis C Virus Ns3/4A Protease the Crystal Structures of Two Protease-Inhibitor Complexes
J.Biol.Chem., 275, 2000
|
|
4F7M
 
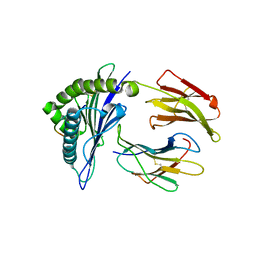 | | Crystal Structure of HLA-A*2402 Complexed with a Newly Identified Peptide from 2009 H1N1 PA (649-658) | | 分子名称: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | 著者 | Liu, J, Zhang, S, Tan, S, Yi, Y, Wu, B, Zhu, F, Wang, H, Qi, J, George, F.G. | | 登録日 | 2012-05-16 | | 公開日 | 2012-10-10 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Cross-Allele Cytotoxic T Lymphocyte Responses against 2009 Pandemic H1N1 Influenza A Virus among HLA-A24 and HLA-A3 Supertype-Positive Individuals.
J.Virol., 86, 2012
|
|
1HBW
 
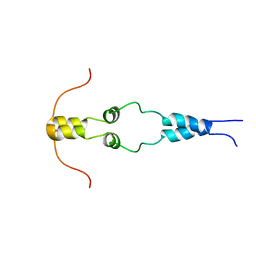 | | Solution nmr structure of the dimerization domain of the yeast transcriptional activator Gal4 (residues 50-106) | | 分子名称: | REGULATORY PROTEIN GAL4 | | 著者 | Hidalgo, P, Ansari, A.Z, Schmidt, P, Hare, B, Simkovic, N, Farrell, S, Shin, E.J, Ptashne, M, Wagner, G. | | 登録日 | 2001-04-20 | | 公開日 | 2001-05-10 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Recruitment of the Transcriptional Machinery Through Gal11P: Structure and Interactions of the GAL4 Dimerization Domain
Genes Dev., 15, 2001
|
|
7U19
 
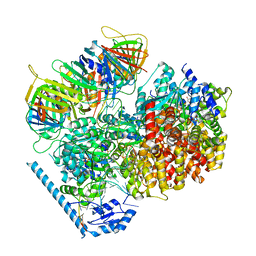 | | RFC:PCNA bound to nicked DNA | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA, MAGNESIUM ION, ... | | 著者 | Liu, X, Gaubitz, C, Pajak, J, Kelch, B.A. | | 登録日 | 2022-02-20 | | 公開日 | 2022-07-06 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | A second DNA binding site on RFC facilitates clamp loading at gapped or nicked DNA.
Elife, 11, 2022
|
|
1JXP
 
 | | BK STRAIN HEPATITIS C VIRUS (HCV) NS3-NS4A | | 分子名称: | NS3 SERINE PROTEASE, NS4A, ZINC ION | | 著者 | Yan, Y, Munshi, S, Chen, Z. | | 登録日 | 1997-08-21 | | 公開日 | 1998-01-14 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Complex of NS3 protease and NS4A peptide of BK strain hepatitis C virus: a 2.2 A resolution structure in a hexagonal crystal form.
Protein Sci., 7, 1998
|
|
8FS5
 
 | | Structure of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 10-nt gapped DNA in step 3 (open 9-1-1 and stably bound chamber DNA) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DDC1 isoform 1, ... | | 著者 | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | 登録日 | 2023-01-09 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.76 Å) | | 主引用文献 | Structures of 9-1-1 DNA checkpoint clamp loading at gaps from start to finish and ramification to biology.
Biorxiv, 2023
|
|
8FS8
 
 | | Structure of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 5-nt gapped DNA (9-1-1 encircling fully bound DNA) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DDC1 isoform 1, ... | | 著者 | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | 登録日 | 2023-01-09 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.04 Å) | | 主引用文献 | Structures of 9-1-1 DNA checkpoint clamp loading at gaps from start to finish and ramification to biology.
Biorxiv, 2023
|
|
8FS7
 
 | | Structure of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 10-nt gapped DNA in step 5 (closed 9-1-1 and stably bound chamber DNA) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DDC1 isoform 1, ... | | 著者 | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | 登録日 | 2023-01-09 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | Structures of 9-1-1 DNA checkpoint clamp loading at gaps from start to finish and ramification to biology.
Biorxiv, 2023
|
|
8FS6
 
 | | Structure of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 10-nt gapped DNA in step 4 (partially closed 9-1-1 and stably bound chamber DNA) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DDC1 isoform 1, ... | | 著者 | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | 登録日 | 2023-01-09 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structures of 9-1-1 DNA checkpoint clamp loading at gaps from start to finish and ramification to biology.
Biorxiv, 2023
|
|
