8HRD
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Delta variant in complex with IMCAS74 Fab and W14 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IMCAS74 Fab heavy chain, IMCAS74 Fab light chain, ... | | 著者 | Zhao, R.C, Wu, L.L, Han, P. | | 登録日 | 2022-12-15 | | 公開日 | 2023-12-20 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.86 Å) | | 主引用文献 | Defining a de novo non-RBM antibody as RBD-8 and its synergistic rescue of immune-evaded antibodies to neutralize Omicron SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5LKA
 
 | | Crystal structure of haloalkane dehalogenase LinB 140A+143L+177W+211L mutant (LinB86) from Sphingobium japonicum UT26 at 1.3 A resolution | | 分子名称: | Haloalkane dehalogenase, THIOCYANATE ION | | 著者 | Degtjarik, O, Rezacova, P, Iermak, I, Chaloupkova, R, Damborsky, J, Kuta Smatanova, I. | | 登録日 | 2016-07-21 | | 公開日 | 2016-10-05 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.298 Å) | | 主引用文献 | Engineering a de novo transport tunnel.
Acs Catalysis, 2016
|
|
5LY1
 
 | | JMJD2A/ KDM4A COMPLEXED WITH NI(II) AND Macrocyclic PEPTIDE Inhibitor CP2 (13-mer) | | 分子名称: | CHLORIDE ION, CP2, GLYCEROL, ... | | 著者 | King, O.N.F, Chowdhury, R, Kawamura, A, Schofield, C.J. | | 登録日 | 2016-09-23 | | 公開日 | 2017-04-12 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Highly selective inhibition of histone demethylases by de novo macrocyclic peptides.
Nat Commun, 8, 2017
|
|
6VDP
 
 | | Crystal structure of SfmD truncated variant | | 分子名称: | 3-methyl-L-tyrosine peroxygenase, HEME C | | 著者 | Shin, I, Liu, A. | | 登録日 | 2019-12-27 | | 公開日 | 2021-03-10 | | 最終更新日 | 2021-07-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A novel catalytic heme cofactor in SfmD with a single thioether bond and a bis -His ligand set revealed by a de novo crystal structural and spectroscopic study.
Chem Sci, 12, 2021
|
|
6VDZ
 
 | |
6VE0
 
 | |
6VDQ
 
 | | Crystal structure of SfmD | | 分子名称: | 3-methyl-L-tyrosine peroxygenase, HEME C | | 著者 | Shin, I, Liu, A. | | 登録日 | 2019-12-27 | | 公開日 | 2021-03-10 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | A novel catalytic heme cofactor in SfmD with a single thioether bond and a bis -His ligand set revealed by a de novo crystal structural and spectroscopic study.
Chem Sci, 12, 2021
|
|
8P6I
 
 | | Crystal structure of the 139H2 Fab fragment bound to Muc1 peptide epitope | | 分子名称: | 139H2 HC, 139H2 LC, Mucin-1 | | 著者 | Beugelink, J.W, Peng, W, Siborova, M, Pronker, M.F, Snijder, J, Janssen, B.J.C. | | 登録日 | 2023-05-26 | | 公開日 | 2024-04-03 | | 最終更新日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Reverse-engineering the anti-MUC1 antibody 139H2 by mass spectrometry-based de novo sequencing.
Life Sci Alliance, 7, 2024
|
|
7BAY
 
 | |
4XTD
 
 | | Structure of the covalent intermediate E-XMP* of the IMP dehydrogenase of Ashbya gossypii | | 分子名称: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase | | 著者 | Buey, R.M, Ledesma-Amaro, R, Balsera, M, de Pereda, J.M, Revuelta, J.L. | | 登録日 | 2015-01-23 | | 公開日 | 2015-07-22 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Increased riboflavin production by manipulation of inosine 5'-monophosphate dehydrogenase in Ashbya gossypii.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4XTI
 
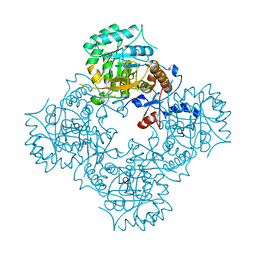 | | Structure of IMP dehydrogenase of Ashbya gossypii with IMP bound to the active site | | 分子名称: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, POTASSIUM ION | | 著者 | Buey, R.M, Ledesma-Amaro, R, Balsera, M, de Pereda, J.M, Revuelta, J.L. | | 登録日 | 2015-01-23 | | 公開日 | 2015-07-22 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Increased riboflavin production by manipulation of inosine 5'-monophosphate dehydrogenase in Ashbya gossypii.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4XWU
 
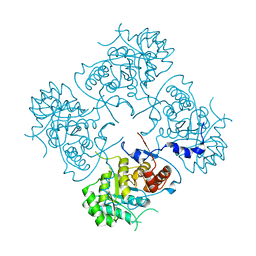 | | Structure of the IMP dehydrogenase from Ashbya gossypii | | 分子名称: | Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase | | 著者 | Buey, R.M, Ledesma-Amaro, R, Balsera, M, de Pereda, J.M, Revuelta, J.L. | | 登録日 | 2015-01-29 | | 公開日 | 2015-07-22 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Increased riboflavin production by manipulation of inosine 5'-monophosphate dehydrogenase in Ashbya gossypii.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4QBQ
 
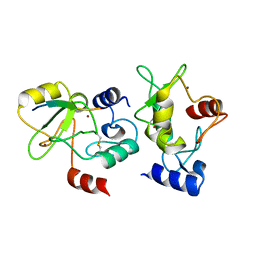 | |
8R12
 
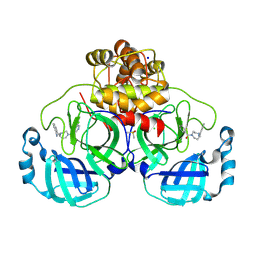 | | Structure of compound 8 bound to SARS-CoV-2 main protease | | 分子名称: | 2-[[4-(5-chloranylpyridin-3-yl)carbonyl-1,4-diazepan-1-yl]methyl]benzenecarbonitrile, 3C-like proteinase, CHLORIDE ION, ... | | 著者 | Mac Sweeney, A, Hazemann, J. | | 登録日 | 2023-11-01 | | 公開日 | 2024-02-07 | | 最終更新日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (1.587 Å) | | 主引用文献 | Identification of SARS-CoV-2 Mpro inhibitors through deep reinforcement learning for de novo drug design and computational chemistry approaches.
Rsc Med Chem, 15, 2024
|
|
8R11
 
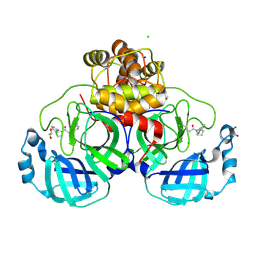 | | Structure of compound 7 bound to SARS-CoV-2 main protease | | 分子名称: | 1,2-ETHANEDIOL, 1-[(2~{S})-2-(3-chlorophenyl)pyrrolidin-1-yl]-2-(5-methylpyridin-3-yl)ethanone, 3C-like proteinase, ... | | 著者 | Mac Sweeney, A, Hazemann, J. | | 登録日 | 2023-11-01 | | 公開日 | 2024-02-07 | | 最終更新日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (1.31 Å) | | 主引用文献 | Identification of SARS-CoV-2 Mpro inhibitors through deep reinforcement learning for de novo drug design and computational chemistry approaches.
Rsc Med Chem, 15, 2024
|
|
8R14
 
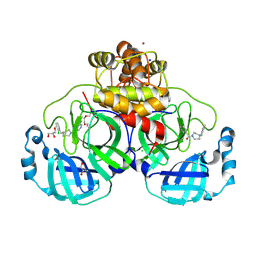 | | Structure of compound 11 bound to SARS-CoV-2 main protease | | 分子名称: | (5-chloranylpyridin-3-yl)-[4-[(2-chlorophenyl)methyl]-1,4-diazepan-1-yl]methanone, 3C-like proteinase, BROMIDE ION, ... | | 著者 | Mac Sweeney, A, Hazemann, J. | | 登録日 | 2023-11-01 | | 公開日 | 2024-02-07 | | 最終更新日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (1.336 Å) | | 主引用文献 | Identification of SARS-CoV-2 Mpro inhibitors through deep reinforcement learning for de novo drug design and computational chemistry approaches.
Rsc Med Chem, 15, 2024
|
|
8R16
 
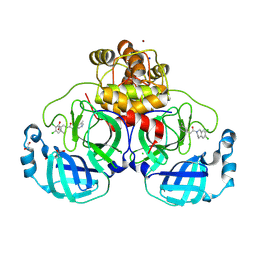 | | Structure of compound 12 bound to SARS-CoV-2 main protease | | 分子名称: | 1,2-ETHANEDIOL, 1-[6,7-bis(chloranyl)-3,4-dihydro-1H-isoquinolin-2-yl]-2-(5-methylpyridin-3-yl)ethanone, 3C-like proteinase, ... | | 著者 | Mac Sweeney, A, Hazemann, J. | | 登録日 | 2023-11-01 | | 公開日 | 2024-02-07 | | 最終更新日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Identification of SARS-CoV-2 Mpro inhibitors through deep reinforcement learning for de novo drug design and computational chemistry approaches.
Rsc Med Chem, 15, 2024
|
|
8QNV
 
 | |
8QNT
 
 | |
4QBS
 
 | |
4QBR
 
 | |
3LXE
 
 | | Human Carbonic Anhydrase I in complex with topiramate | | 分子名称: | Carbonic anhydrase 1, ZINC ION, [(3aS,5aR,8aR,8bS)-2,2,7,7-tetramethyltetrahydro-3aH-bis[1,3]dioxolo[4,5-b:4',5'-d]pyran-3a-yl]methyl sulfamate | | 著者 | Alterio, V, De Simone, G, Monti, S.M, Truppo, E. | | 登録日 | 2010-02-25 | | 公開日 | 2010-07-21 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The first example of a significant active site conformational rearrangement in a carbonic anhydrase-inhibitor adduct: the carbonic anhydrase I-topiramate complex.
Org.Biomol.Chem., 2010
|
|
8ATV
 
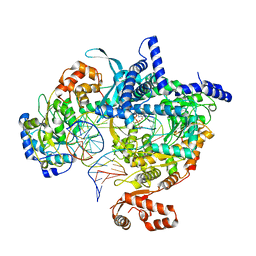 | | Cryo-EM structure of yeast mitochondrial RNA polymerase transcription initiation complex with 5-mer RNA, pppGpGpApApA (IC5) | | 分子名称: | DNA (36-MER), DNA-directed RNA polymerase, mitochondrial, ... | | 著者 | Goovaerts, Q, Shen, J, Patel, S.S, Das, K. | | 登録日 | 2022-08-24 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-11-01 | | 実験手法 | ELECTRON MICROSCOPY (3.39 Å) | | 主引用文献 | Structures illustrate step-by-step mitochondrial transcription initiation.
Nature, 622, 2023
|
|
8ATW
 
 | | Cryo-EM structure of yeast mitochondrial RNA polymerase transcription initiation complex with 6-mer RNA, pppGpGpApApApU (IC6) | | 分子名称: | DNA-directed RNA polymerase, mitochondrial, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Goovaerts, Q, Shen, J, Patel, S.S, Das, K. | | 登録日 | 2022-08-24 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-11-01 | | 実験手法 | ELECTRON MICROSCOPY (3.62 Å) | | 主引用文献 | Structures illustrate step-by-step mitochondrial transcription initiation.
Nature, 622, 2023
|
|
8ATT
 
 | | Cryo-EM structure of yeast mitochondrial RNA polymerase transcription initiation complex with 4-mer RNA, pppGpGpUpA (IC4) | | 分子名称: | DNA-directed RNA polymerase, mitochondrial, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Goovaerts, Q, Shen, J, Patel, S.S, Das, K. | | 登録日 | 2022-08-24 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-11-01 | | 実験手法 | ELECTRON MICROSCOPY (3.44 Å) | | 主引用文献 | Structures illustrate step-by-step mitochondrial transcription initiation.
Nature, 622, 2023
|
|
