8SMZ
 
 | | Cryo-EM structure of the human nucleosome core particle in complex with RNF168 and UbcH5c~Ub (UbcH5c chemically conjugated to histone H2A. No density for Ub.) (Class 4) | | 分子名称: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | 著者 | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | 登録日 | 2023-04-26 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8SN0
 
 | | Cryo-EM structure of the human nucleosome core particle in complex with RNF168 and UbcH5c~Ub (UbcH5c chemically conjugated to histone H2A. No density for Ub.) (class 5) | | 分子名称: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | 著者 | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | 登録日 | 2023-04-26 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8SN1
 
 | | Cryo-EM structure of the human nucleosome core particle in complex with RNF168 and UbcH5c~Ub (UbcH5c chemically conjugated to histone H2A. No density for Ub.) (class 6) | | 分子名称: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | 著者 | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | 登録日 | 2023-04-26 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
4GK0
 
 | | Crystal structure of human Rev3-Rev7-Rev1 complex | | 分子名称: | DNA polymerase zeta catalytic subunit, DNA repair protein REV1, Mitotic spindle assembly checkpoint protein MAD2B | | 著者 | Tao, J, Min, X, Wei, X. | | 登録日 | 2012-08-10 | | 公開日 | 2013-03-13 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural insights into the assembly of human translesion polymerase complexes
Protein Cell, 3, 2012
|
|
8TXX
 
 | | Cryo-EM structure of the human nucleosome core particle ubiquitylated at histone H2A K15 in complex with RNF168 (Class 3) | | 分子名称: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | 著者 | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | 登録日 | 2023-08-24 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8TXV
 
 | | Cryo-EM structure of the human nucleosome core particle ubiquitylated at histone H2A K15 in complex with RNF168 (Class 1) | | 分子名称: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | 著者 | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | 登録日 | 2023-08-24 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8TXW
 
 | | Cryo-EM structure of the human nucleosome core particle ubiquitylated at histone H2A K15 in complex with RNF168 (Class 2) | | 分子名称: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | 著者 | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | 登録日 | 2023-08-24 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8JNF
 
 | | The cryo-EM structure of the RAD51 filament bound to the nucleosome | | 分子名称: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-06-06 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (6.91 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8H1T
 
 | | Cryo-EM structure of BAP1-ASXL1 bound to chromatosome | | 分子名称: | DNA (187-MER), Histone H1.4, Histone H2A type 1-D, ... | | 著者 | Ge, W, Yu, C, Xu, R.M. | | 登録日 | 2022-10-04 | | 公開日 | 2023-02-01 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Basis of the H2AK119 specificity of the Polycomb repressive deubiquitinase.
Nature, 616, 2023
|
|
8TNP
 
 | | Cryo-EM structure of DDB1dB:CRBN:Pomalidomide:SD40 | | 分子名称: | DNA damage-binding protein 1, Maltose/maltodextrin-binding periplasmic protein,SD40, Protein cereblon, ... | | 著者 | Roy Burman, S.S, Hunkeler, M, Fischer, E.S. | | 登録日 | 2023-08-02 | | 公開日 | 2024-03-13 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Continuous evolution of compact protein degradation tags regulated by selective molecular glues.
Science, 383, 2024
|
|
2HTF
 
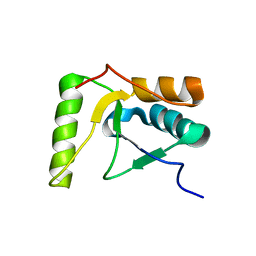 | | The solution structure of the BRCT domain from human polymerase reveals homology with the TdT BRCT domain | | 分子名称: | DNA polymerase mu | | 著者 | DeRose, E.F, Clarkson, M.W, Gilmore, S.A, Ramsden, D.A, Mueller, G.A, London, R.E, Lee, A.L. | | 登録日 | 2006-07-25 | | 公開日 | 2007-02-27 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of polymerase mu's BRCT Domain reveals an element essential for its role in nonhomologous end joining.
Biochemistry, 46, 2007
|
|
1V4A
 
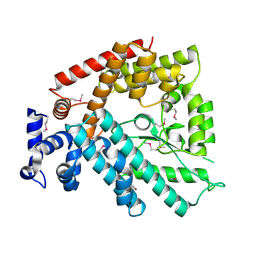 | | Structure of the N-terminal Domain of Escherichia coli Glutamine Synthetase adenylyltransferase | | 分子名称: | Glutamate-ammonia-ligase adenylyltransferase | | 著者 | Xu, Y, Zhang, R, Joachimiak, A, Carr, P.D, Ollis, D.L, Vasudevan, S.G. | | 登録日 | 2003-11-12 | | 公開日 | 2004-07-27 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of the n-terminal domain of Escherichia coli glutamine synthetase adenylyltransferase
Structure, 12, 2004
|
|
7EJE
 
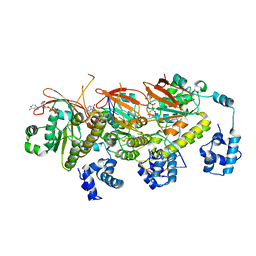 | | human RAD51 post-synaptic complex | | 分子名称: | DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA repair protein RAD51 homolog 1, ... | | 著者 | Zhao, L.Y, Xu, J.F, Wang, H.W. | | 登録日 | 2021-04-02 | | 公開日 | 2022-03-16 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.98 Å) | | 主引用文献 | Mechanisms of distinctive mismatch tolerance between Rad51 and Dmc1 in homologous recombination.
Nucleic Acids Res., 49, 2021
|
|
8GUK
 
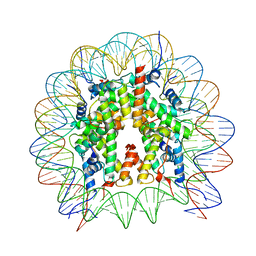 | | Human nucleosome core particle (free form) | | 分子名称: | DNA (147-mer), Histone H2A type 1, Histone H2B type 1-J, ... | | 著者 | Onishi, S, Sato, K, Nishizawa, T, Nureki, O, Ogata, K, Sengoku, T. | | 登録日 | 2022-09-12 | | 公開日 | 2023-09-20 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (2.51 Å) | | 主引用文献 | Structure of the human Bre1 complex bound to the nucleosome.
Nat Commun, 15, 2024
|
|
7LYA
 
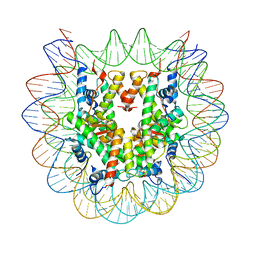 | | Cryo-EM structure of the human nucleosome core particle with linked histone proteins H2A and H2B | | 分子名称: | DNA (146-MER), DNA (147-MER), Histone H2A type 1-B/E, ... | | 著者 | Hu, Q, Botuyan, M.V, Zhao, D, Cui, D, Mer, E, Mer, G. | | 登録日 | 2021-03-06 | | 公開日 | 2021-07-28 | | 最終更新日 | 2021-09-01 | | 実験手法 | ELECTRON MICROSCOPY (2.91 Å) | | 主引用文献 | Mechanisms of BRCA1-BARD1 nucleosome recognition and ubiquitylation.
Nature, 596, 2021
|
|
8U13
 
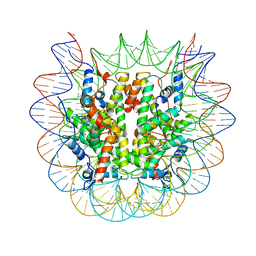 | | Cryo-EM structure of the human nucleosome core particle ubiquitylated at histone H2A lysine 15 in complex with RNF168-UbcH5c (class 1) | | 分子名称: | DNA (146-MER), DNA (147-MER), E3 ubiquitin-protein ligase RNF168, ... | | 著者 | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | 登録日 | 2023-08-30 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
7YQH
 
 | | Cryo-EM structure of 8-subunit Smc5/6 | | 分子名称: | DNA repair protein KRE29, E3 SUMO-protein ligase MMS21, Non-structural maintenance of chromosome element 3, ... | | 著者 | Qian, L, Jun, Z, Xiang, Z, Tong, C, Zhaoning, W, Duo, J, Zhenguo, C, Lanfeng, W. | | 登録日 | 2022-08-07 | | 公開日 | 2024-01-31 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (5.6 Å) | | 主引用文献 | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
8I4V
 
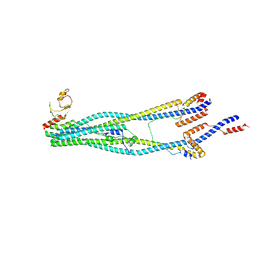 | | Cryo-EM structure of 5-subunit Smc5/6 arm region | | 分子名称: | DNA repair protein KRE29, E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, ... | | 著者 | Qian, L, Jun, Z, Xiang, Z, Cheng, T, Zhaoning, W, Zhenguo, C, Wang, L. | | 登録日 | 2023-01-21 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (5.97 Å) | | 主引用文献 | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
3UJ3
 
 | |
3T79
 
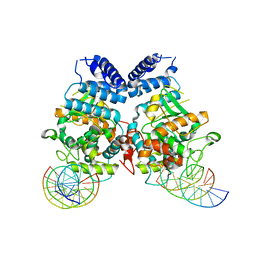 | | Ndc10: a platform for inner kinetochore assembly in budding yeast | | 分子名称: | DNA (5'-D(P*AP*AP*AP*AP*AP*TP*TP*TP*TP*AP*TP*AP*AP*AP*T)-3'), DNA (5'-D(P*AP*AP*AP*TP*TP*TP*TP*AP*TP*AP*AP*AP*TP*TP*A)-3'), DNA (5'-D(P*TP*AP*AP*TP*TP*TP*AP*TP*AP*AP*AP*AP*TP*T)-3'), ... | | 著者 | Cho, U.S, Harrison, S.C. | | 登録日 | 2011-07-29 | | 公開日 | 2011-12-07 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3.6113 Å) | | 主引用文献 | Ndc10 is a platform for inner kinetochore assembly in budding yeast.
Nat.Struct.Mol.Biol., 19, 2011
|
|
6L8N
 
 | | Crystal structure of the K. lactis Rad5 | | 分子名称: | DNA repair protein RAD5, ZINC ION | | 著者 | Shen, M, Xiang, S. | | 登録日 | 2019-11-06 | | 公開日 | 2020-11-11 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Structural basis for the multi-activity factor Rad5 in replication stress tolerance.
Nat Commun, 12, 2021
|
|
5V3O
 
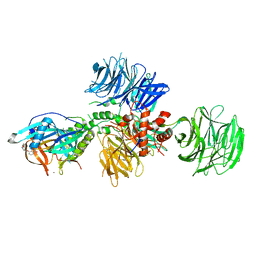 | | Cereblon in complex with DDB1 and CC-220 | | 分子名称: | (3S)-3-[4-({4-[(morpholin-4-yl)methyl]phenyl}methoxy)-1-oxo-1,3-dihydro-2H-isoindol-2-yl]piperidine-2,6-dione, DNA damage-binding protein 1, Protein cereblon, ... | | 著者 | Matyskiela, M, Pagarigan, B, Chamberlain, P. | | 登録日 | 2017-03-07 | | 公開日 | 2017-05-03 | | 最終更新日 | 2018-02-07 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | A Cereblon Modulator (CC-220) with Improved Degradation of Ikaros and Aiolos.
J. Med. Chem., 61, 2018
|
|
6Q0W
 
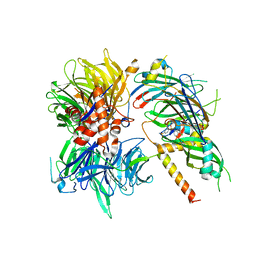 | | Structure of DDB1-DDA1-DCAF15 complex bound to Indisulam and RBM39 | | 分子名称: | DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, DNA damage-binding protein 1, ... | | 著者 | Faust, T, Yoon, H, Nowak, R.P, Donovan, K.A, Li, Z, Cai, Q, Eleuteri, N.A, Zhang, T, Gray, N.S, Fischer, E.S. | | 登録日 | 2019-08-02 | | 公開日 | 2019-11-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural complementarity facilitates E7820-mediated degradation of RBM39 by DCAF15.
Nat.Chem.Biol., 16, 2020
|
|
6Q0R
 
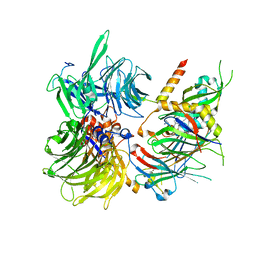 | | Structure of DDB1-DDA1-DCAF15 complex bound to E7820 and RBM39 | | 分子名称: | 3-cyano-N-(3-cyano-4-methyl-1H-indol-7-yl)benzene-1-sulfonamide, DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, ... | | 著者 | Faust, T, Yoon, H, Nowak, R.P, Donovan, K.A, Li, Z, Cai, Q, Eleuteri, N.A, Zhang, T, Gray, N.S, Fischer, E.S. | | 登録日 | 2019-08-02 | | 公開日 | 2019-11-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural complementarity facilitates E7820-mediated degradation of RBM39 by DCAF15.
Nat.Chem.Biol., 16, 2020
|
|
7ZT6
 
 | |
