4XTI
 
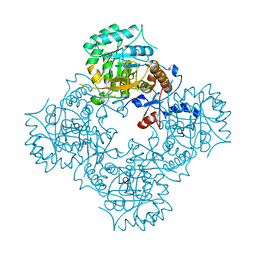 | | Structure of IMP dehydrogenase of Ashbya gossypii with IMP bound to the active site | | 分子名称: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, POTASSIUM ION | | 著者 | Buey, R.M, Ledesma-Amaro, R, Balsera, M, de Pereda, J.M, Revuelta, J.L. | | 登録日 | 2015-01-23 | | 公開日 | 2015-07-22 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Increased riboflavin production by manipulation of inosine 5'-monophosphate dehydrogenase in Ashbya gossypii.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4XWU
 
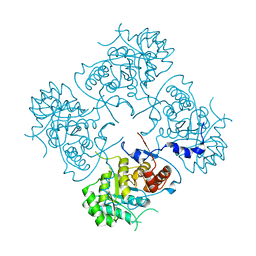 | | Structure of the IMP dehydrogenase from Ashbya gossypii | | 分子名称: | Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase | | 著者 | Buey, R.M, Ledesma-Amaro, R, Balsera, M, de Pereda, J.M, Revuelta, J.L. | | 登録日 | 2015-01-29 | | 公開日 | 2015-07-22 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Increased riboflavin production by manipulation of inosine 5'-monophosphate dehydrogenase in Ashbya gossypii.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
6LDF
 
 | |
6LDE
 
 | |
6LDG
 
 | |
8QNV
 
 | |
8QNT
 
 | |
8R11
 
 | | Structure of compound 7 bound to SARS-CoV-2 main protease | | 分子名称: | 1,2-ETHANEDIOL, 1-[(2~{S})-2-(3-chlorophenyl)pyrrolidin-1-yl]-2-(5-methylpyridin-3-yl)ethanone, 3C-like proteinase, ... | | 著者 | Mac Sweeney, A, Hazemann, J. | | 登録日 | 2023-11-01 | | 公開日 | 2024-02-07 | | 最終更新日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (1.31 Å) | | 主引用文献 | Identification of SARS-CoV-2 Mpro inhibitors through deep reinforcement learning for de novo drug design and computational chemistry approaches.
Rsc Med Chem, 15, 2024
|
|
8R14
 
 | | Structure of compound 11 bound to SARS-CoV-2 main protease | | 分子名称: | (5-chloranylpyridin-3-yl)-[4-[(2-chlorophenyl)methyl]-1,4-diazepan-1-yl]methanone, 3C-like proteinase, BROMIDE ION, ... | | 著者 | Mac Sweeney, A, Hazemann, J. | | 登録日 | 2023-11-01 | | 公開日 | 2024-02-07 | | 最終更新日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (1.336 Å) | | 主引用文献 | Identification of SARS-CoV-2 Mpro inhibitors through deep reinforcement learning for de novo drug design and computational chemistry approaches.
Rsc Med Chem, 15, 2024
|
|
8R16
 
 | | Structure of compound 12 bound to SARS-CoV-2 main protease | | 分子名称: | 1,2-ETHANEDIOL, 1-[6,7-bis(chloranyl)-3,4-dihydro-1H-isoquinolin-2-yl]-2-(5-methylpyridin-3-yl)ethanone, 3C-like proteinase, ... | | 著者 | Mac Sweeney, A, Hazemann, J. | | 登録日 | 2023-11-01 | | 公開日 | 2024-02-07 | | 最終更新日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Identification of SARS-CoV-2 Mpro inhibitors through deep reinforcement learning for de novo drug design and computational chemistry approaches.
Rsc Med Chem, 15, 2024
|
|
8R12
 
 | | Structure of compound 8 bound to SARS-CoV-2 main protease | | 分子名称: | 2-[[4-(5-chloranylpyridin-3-yl)carbonyl-1,4-diazepan-1-yl]methyl]benzenecarbonitrile, 3C-like proteinase, CHLORIDE ION, ... | | 著者 | Mac Sweeney, A, Hazemann, J. | | 登録日 | 2023-11-01 | | 公開日 | 2024-02-07 | | 最終更新日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (1.587 Å) | | 主引用文献 | Identification of SARS-CoV-2 Mpro inhibitors through deep reinforcement learning for de novo drug design and computational chemistry approaches.
Rsc Med Chem, 15, 2024
|
|
4QBS
 
 | |
4QBR
 
 | |
4QBQ
 
 | |
6U91
 
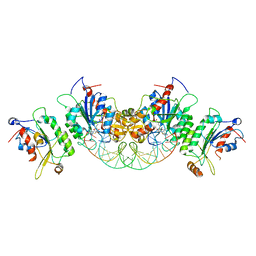 | | Crystal structure of DNMT3B(Q772R)-DNMT3L in complex with CpGpT DNA | | 分子名称: | CpGpT DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B, ... | | 著者 | Gao, L, Song, J. | | 登録日 | 2019-09-06 | | 公開日 | 2020-06-10 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.99998879 Å) | | 主引用文献 | Comprehensive structure-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms
To Be Published
|
|
6U90
 
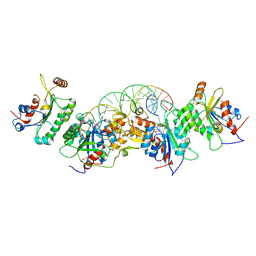 | | Crystal structure of DNMT3B(N779A)-DNMT3L in complex with CpGpT DNA | | 分子名称: | CpGpT DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B, ... | | 著者 | Gao, L, Song, J. | | 登録日 | 2019-09-06 | | 公開日 | 2020-06-10 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.00081754 Å) | | 主引用文献 | Comprehensive structure-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms
To Be Published
|
|
6VDP
 
 | | Crystal structure of SfmD truncated variant | | 分子名称: | 3-methyl-L-tyrosine peroxygenase, HEME C | | 著者 | Shin, I, Liu, A. | | 登録日 | 2019-12-27 | | 公開日 | 2021-03-10 | | 最終更新日 | 2021-07-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A novel catalytic heme cofactor in SfmD with a single thioether bond and a bis -His ligand set revealed by a de novo crystal structural and spectroscopic study.
Chem Sci, 12, 2021
|
|
6VDZ
 
 | |
6VE0
 
 | |
6VDQ
 
 | | Crystal structure of SfmD | | 分子名称: | 3-methyl-L-tyrosine peroxygenase, HEME C | | 著者 | Shin, I, Liu, A. | | 登録日 | 2019-12-27 | | 公開日 | 2021-03-10 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | A novel catalytic heme cofactor in SfmD with a single thioether bond and a bis -His ligand set revealed by a de novo crystal structural and spectroscopic study.
Chem Sci, 12, 2021
|
|
3LXE
 
 | | Human Carbonic Anhydrase I in complex with topiramate | | 分子名称: | Carbonic anhydrase 1, ZINC ION, [(3aS,5aR,8aR,8bS)-2,2,7,7-tetramethyltetrahydro-3aH-bis[1,3]dioxolo[4,5-b:4',5'-d]pyran-3a-yl]methyl sulfamate | | 著者 | Alterio, V, De Simone, G, Monti, S.M, Truppo, E. | | 登録日 | 2010-02-25 | | 公開日 | 2010-07-21 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The first example of a significant active site conformational rearrangement in a carbonic anhydrase-inhibitor adduct: the carbonic anhydrase I-topiramate complex.
Org.Biomol.Chem., 2010
|
|
4IGH
 
 | | High resolution crystal structure of human dihydroorotate dehydrogenase bound with 4-quinoline carboxylic acid analog | | 分子名称: | 3-[decyl(dimethyl)ammonio]propane-1-sulfonate, 6-fluoro-2-[2-methyl-4-phenoxy-5-(propan-2-yl)phenyl]quinoline-4-carboxylic acid, Dihydroorotate dehydrogenase (quinone), ... | | 著者 | Deng, X, Das, P, Fontoura, B.M.A, Phillips, M.A, De Brabander, J.K. | | 登録日 | 2012-12-17 | | 公開日 | 2013-08-21 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | SAR Based Optimization of a 4-Quinoline Carboxylic Acid Analog with Potent Anti-Viral Activity.
ACS MED.CHEM.LETT., 4, 2013
|
|
8UJA
 
 | |
6EBS
 
 | | Crystal structure of Leishmania major dihydroorotate dehydrogenase mutant H174A in complex with orotate | | 分子名称: | Dihydroorotate dehydrogenase (fumarate), FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | 著者 | Reis, R.A.G, Pinheiro, M.P, de Souza, A.L, Hunter, W.N, Nonato, M.C. | | 登録日 | 2018-08-07 | | 公開日 | 2019-08-21 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Crystal structure of Leishmania major dihydroorotate dehydrogenase mutant H174A in complex with orotate
To Be Published
|
|
7JMH
 
 | | Functional Pathways of Biomolecules Retrieved from Single-particle Snapshots - Frame 35 - State 4 (S4) | | 分子名称: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, ZINC ION, ... | | 著者 | Dashti, A, des Georges, A, Frank, J, Ourmazd, A. | | 登録日 | 2020-07-31 | | 公開日 | 2020-08-12 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Retrieving functional pathways of biomolecules from single-particle snapshots.
Nat Commun, 11, 2020
|
|
