7Z5I
 
 | |
7Z5K
 
 | |
8G32
 
 | | Pro-form of a CDCL short from E. anophelis | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | 著者 | Johnstone, B.A, Christie, M.P, Morton, C.J, Parker, M.W. | | 登録日 | 2023-02-06 | | 公開日 | 2024-02-07 | | 最終更新日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Distant relatives of a eukaryotic cell-specific toxin family evolved a complement-like mechanism to kill bacteria.
Nat Commun, 15, 2024
|
|
8GF4
 
 | |
7BIN
 
 | | Salmonella export gate and rod refined in focussed C1 map | | 分子名称: | Flagellar basal body rod protein FlgB, Flagellar basal-body rod protein FlgC, Flagellar basal-body rod protein FlgF, ... | | 著者 | Johnson, S, Furlong, E, Lea, S.M. | | 登録日 | 2021-01-12 | | 公開日 | 2021-05-05 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Molecular structure of the intact bacterial flagellar basal body.
Nat Microbiol, 6, 2021
|
|
6Z0M
 
 | | Het-Ncap - De novo designed three-helix heterodimer with Cysteine at the Ncap position of the alpha-helix | | 分子名称: | Cys-Ncap strand, Positive Strand, SULFATE ION, ... | | 著者 | McEwen, A.G, Poussin-Courmontagne, P, Naudin, E.A, DeGrado, W.F, Torbeev, V. | | 登録日 | 2020-05-09 | | 公開日 | 2021-03-17 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Acyl Transfer Catalytic Activity in De Novo Designed Protein with N-Terminus of alpha-Helix As Oxyanion-Binding Site.
J.Am.Chem.Soc., 143, 2021
|
|
8GDW
 
 | |
8U9F
 
 | | Crystal structure of Bacteroides thetaiotamicron BT1285 in complex with NaI | | 分子名称: | 1,2-ETHANEDIOL, Endo-beta-N-acetylglucosaminidase, IODIDE ION, ... | | 著者 | Sastre, D.E, Navarro, M.V.A.S, Sundberg, E.J. | | 登録日 | 2023-09-19 | | 公開日 | 2024-05-29 | | 最終更新日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (1.08 Å) | | 主引用文献 | Human gut microbes express functionally distinct endoglycosidases to metabolize the same N-glycan substrate.
Nat Commun, 15, 2024
|
|
8GKL
 
 | |
6QG9
 
 | | Crystal structure of Ideonella sakaiensis MHETase | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | 著者 | Palm, G.J, Reisky, L, Boettcher, D, Mueller, H, Michels, E.A.P, Walczak, C, Berndt, L, Weiss, M.S, Bornscheuer, U.T, Weber, G. | | 登録日 | 2019-01-10 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate.
Nat Commun, 10, 2019
|
|
5LVP
 
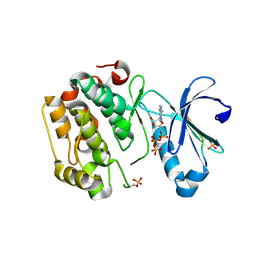 | | Human PDK1 Kinase Domain in Complex with an HM-Peptide Bound to the PIF-Pocket | | 分子名称: | 3-phosphoinositide-dependent protein kinase 1, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | 著者 | Schulze, J.O, Saladino, G, Busschots, K, Neimanis, S, Suess, E, Odadzic, D, Zeuzem, S, Hindie, V, Herbrand, A.K, Lisa, M.N, Alzari, P.M, Gervasio, F.L, Biondi, R.M. | | 登録日 | 2016-09-14 | | 公開日 | 2016-10-19 | | 最終更新日 | 2019-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Bidirectional Allosteric Communication between the ATP-Binding Site and the Regulatory PIF Pocket in PDK1 Protein Kinase.
Cell Chem Biol, 23, 2016
|
|
6ZHH
 
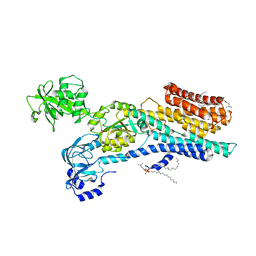 | | Ca2+-ATPase from Listeria Monocytogenes with G4 insertion. | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, BERYLLIUM TRIFLUORIDE ION, Calcium-transporting ATPase, ... | | 著者 | Basse Hansen, S, Dyla, M, Neumann, C, Quistgaard, E.M.H, Lauwring Andersen, J, Kjaergaard, M, Nissen, P. | | 登録日 | 2020-06-23 | | 公開日 | 2021-05-19 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The Crystal Structure of the Ca 2+ -ATPase 1 from Listeria monocytogenes reveals a Pump Primed for Dephosphorylation.
J.Mol.Biol., 433, 2021
|
|
7B91
 
 | |
5O56
 
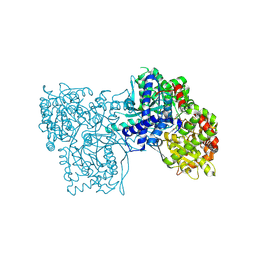 | | Glycogen Phosphorylase b in complex with 29b | | 分子名称: | (2~{R},3~{S},4~{R},5~{R},6~{R})-5-azanyl-2-(hydroxymethyl)-6-(3-naphthalen-2-yl-1~{H}-1,2,4-triazol-5-yl)oxane-3,4-diol, Glycogen phosphorylase, muscle form, ... | | 著者 | Solovou, T.G.A, Kyriakis, E, Stravodimos, G.A, Kantsadi, A.L, Chatzileontiadou, D.S, Skamnaki, V.T, Leonidas, D.D. | | 登録日 | 2017-05-31 | | 公開日 | 2017-09-27 | | 最終更新日 | 2017-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Nanomolar Inhibitors of Glycogen Phosphorylase Based on beta-d-Glucosaminyl Heterocycles: A Combined Synthetic, Enzyme Kinetic, and Protein Crystallography Study.
J. Med. Chem., 60, 2017
|
|
8QT5
 
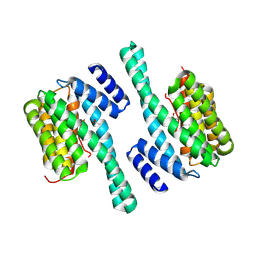 | |
4YO5
 
 | | EAEC T6SS TssA-Cterminus | | 分子名称: | TssA | | 著者 | Durand, E, Zoued, A, Spinelli, S, Douzi, B, Brunet, Y.R, Bebeacua, C, Legrand, P, Journet, L, Mignot, T, Cambillau, C, Cascales, E. | | 登録日 | 2015-03-11 | | 公開日 | 2016-02-17 | | 最終更新日 | 2017-06-14 | | 実験手法 | X-RAY DIFFRACTION (3.35 Å) | | 主引用文献 | Priming and polymerization of a bacterial contractile tail structure.
Nature, 531, 2016
|
|
6FQS
 
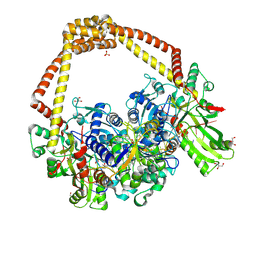 | | 3.11A complex of S.Aureus gyrase with imidazopyrazinone T3 and DNA | | 分子名称: | 5-cyclopropyl-8-fluoranyl-7-pyridin-4-yl-imidazo[1,2-a]quinoxalin-4-one, DNA (5'-D(*GP*AP*GP*AP*GP*TP*AP*T*GP*GP*CP*CP*AP*TP*AP*CP*TP*CP*TP*T)-3'), DNA gyrase subunit A, ... | | 著者 | Bax, B.D, Germe, T, Basque, E, Maxwell, A. | | 登録日 | 2018-02-14 | | 公開日 | 2018-04-04 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.11 Å) | | 主引用文献 | A new class of antibacterials, the imidazopyrazinones, reveal structural transitions involved in DNA gyrase poisoning and mechanisms of resistance.
Nucleic Acids Res., 46, 2018
|
|
1H1R
 
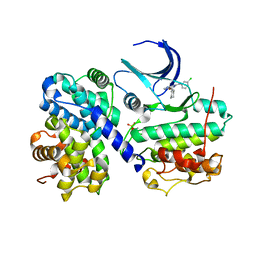 | | Structure of human Thr160-phospho CDK2/cyclin A complexed with the inhibitor NU6086 | | 分子名称: | 6-CYCLOHEXYLMETHOXY-2-(3'-CHLOROANILINO) PURINE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2 | | 著者 | Davies, T.G, Noble, M.E.M, Endicott, J.A, Johnson, L.N. | | 登録日 | 2002-07-21 | | 公開日 | 2002-09-19 | | 最終更新日 | 2019-10-02 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-based design of a potent purine-based cyclin-dependent kinase inhibitor.
Nat. Struct. Biol., 9, 2002
|
|
5OCO
 
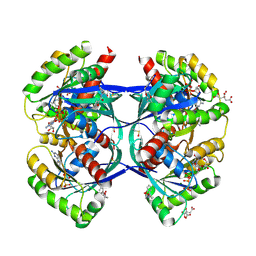 | | Discovery of small molecules binding to KRAS via high affinity antibody fragment competition method. | | 分子名称: | CITRIC ACID, DI(HYDROXYETHYL)ETHER, GTPase KRas, ... | | 著者 | Cruz-Migoni, A, Ehebauer, M.T, Phillips, S.E.V, Quevedo, C.E, Rabbitts, T.H. | | 登録日 | 2017-07-03 | | 公開日 | 2018-08-22 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Small molecule inhibitors of RAS-effector protein interactions derived using an intracellular antibody fragment.
Nat Commun, 9, 2018
|
|
4YO3
 
 | | Enteroaggregative Escherichia Coli TssA N-terminal fragment | | 分子名称: | TssA | | 著者 | Durand, E, Zoued, A, Spinelli, S, Douzi, B, Brunet, Y.R, Bebeacua, C, Legrand, P, Journet, L, Mignot, T, Cambillau, C, Cascales, E. | | 登録日 | 2015-03-11 | | 公開日 | 2016-02-17 | | 最終更新日 | 2017-08-30 | | 実験手法 | X-RAY DIFFRACTION (3.37 Å) | | 主引用文献 | Priming and polymerization of a bacterial contractile tail structure.
Nature, 531, 2016
|
|
7UN0
 
 | | Structure of MAP kinase phosphatase 5 in complex with 3,3-dimethyl-1-((9-chloro-5,6-dihydrobenzo[h]quinazolin-2-yl)thio)butan-2-one, an allosteric inhibitor | | 分子名称: | 1-[(9-chlorobenzo[h]quinazolin-2-yl)sulfanyl]-3,3-dimethylbutan-2-one, Dual specificity protein phosphatase 10 | | 著者 | Gannam, Z.T.K, Jamali, H, Lolis, E, Anderson, K.S, Ellman, J.A, Bennett, A.M. | | 登録日 | 2022-04-08 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Defining the structure-activity relationship for a novel class of allosteric MKP5 inhibitors.
Eur.J.Med.Chem., 243, 2022
|
|
1HWY
 
 | | BOVINE GLUTAMATE DEHYDROGENASE COMPLEXED WITH NAD AND 2-OXOGLUTARATE | | 分子名称: | 2-OXOGLUTARIC ACID, GLUTAMATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Smith, T.J, Peterson, P.E, Schmidt, T, Fang, J, Stanley, C.A. | | 登録日 | 2001-01-10 | | 公開日 | 2001-01-31 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structures of bovine glutamate dehydrogenase complexes elucidate the mechanism of purine regulation.
J.Mol.Biol., 307, 2001
|
|
7GB8
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with JAN-GHE-83b26c96-14 (Mpro-x10247) | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(4-methylpyridin-3-yl)-2-[3-(trifluoromethyl)phenyl]acetamide | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.957 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
6AMO
 
 | | STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) TERNARY COMPLEX WITH A DOUBLE STRANDED DNA AND AN INCOMING D4TTP AT PH 7.0 | | 分子名称: | 2',3'-DEHYDRO-2',3'-DEOXY-THYMIDINE 5'-TRIPHOSPHATE, DNA (27-MER), DNA (5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*(DDG))-3'), ... | | 著者 | Martinez, S.E, Das, K, Arnold, E. | | 登録日 | 2017-08-10 | | 公開日 | 2018-08-15 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.497 Å) | | 主引用文献 | Structure of HIV-1 reverse transcriptase/d4TTP complex: Novel DNA cross-linking site and pH-dependent conformational changes.
Protein Sci., 28, 2019
|
|
7GBK
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ALP-POS-95b75b4d-2 (Mpro-x10359) | | 分子名称: | 2-(3-hydroxyphenyl)-N-(4-methylpyridin-3-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
