4FWT
 
 | |
6P1H
 
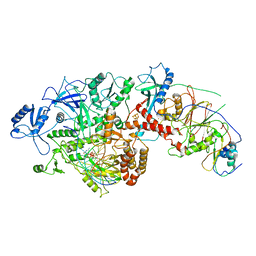 | | Cryo-EM Structure of DNA Polymerase Delta Holoenzyme | | 分子名称: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (30-MER), ... | | 著者 | Jain, R, Rice, W, Aggarwal, A.K. | | 登録日 | 2019-05-19 | | 公開日 | 2019-10-02 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structure and dynamics of eukaryotic DNA polymerase delta holoenzyme.
Nat.Struct.Mol.Biol., 26, 2019
|
|
4FRG
 
 | | Crystal structure of the cobalamin riboswitch aptamer domain | | 分子名称: | Hydroxocobalamin, IRIDIUM (III) ION, MAGNESIUM ION, ... | | 著者 | Reyes, F.E, Johnson, J.E, Polaski, J.T, Batey, R.T. | | 登録日 | 2012-06-26 | | 公開日 | 2012-10-17 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | B12 cofactors directly stabilize an mRNA regulatory switch.
Nature, 492, 2012
|
|
3Q36
 
 | |
8KG9
 
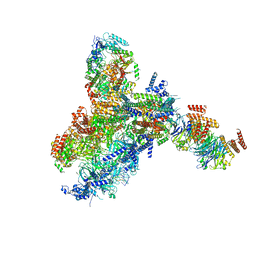 | | Yeast replisome in state III | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45, DNA (61-mer), ... | | 著者 | Dang, S, Zhai, Y, Feng, J, Yu, D, Xu, Z. | | 登録日 | 2023-08-17 | | 公開日 | 2023-12-06 | | 実験手法 | ELECTRON MICROSCOPY (4.52 Å) | | 主引用文献 | Synergism between CMG helicase and leading strand DNA polymerase at replication fork.
Nat Commun, 14, 2023
|
|
2LTO
 
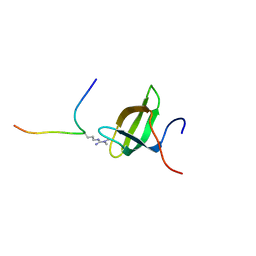 | | TDRD3 complex | | 分子名称: | DNA-directed RNA polymerase II subunit RPB1, Tudor domain-containing protein 3 | | 著者 | Sikorsky, T. | | 登録日 | 2012-05-30 | | 公開日 | 2013-01-16 | | 最終更新日 | 2023-11-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Recognition of asymmetrically dimethylated arginine by TDRD3.
Nucleic Acids Res., 40, 2012
|
|
6XQB
 
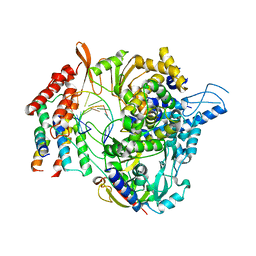 | | SARS-CoV-2 RdRp/RNA complex | | 分子名称: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | 著者 | Liu, B, Shi, W, Yang, Y. | | 登録日 | 2020-07-09 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure of SARS-CoV-2 RdRp/RNA complex at 3.4 Angstroms resolution
To Be Published
|
|
6VEM
 
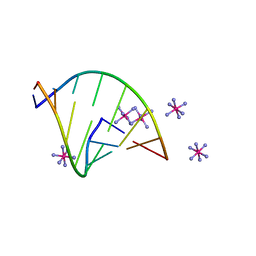 | | Structure of RNA octamer | | 分子名称: | COBALT HEXAMMINE(III), Modified Octamer RNA | | 著者 | Pallan, P.S, Egli, M. | | 登録日 | 2020-01-02 | | 公開日 | 2020-11-18 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Synthesis, chirality-dependent conformational and biological properties of siRNAs containing 5'-(R)- and 5'-(S)-C-methyl-guanosine.
Nucleic Acids Res., 48, 2020
|
|
4R71
 
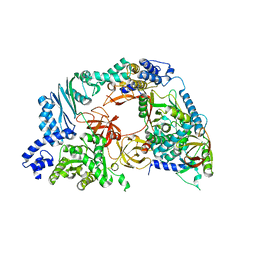 | | Structure of the Qbeta holoenzyme complex in the P1211 crystal form | | 分子名称: | 30S ribosomal protein S1, Elongation factor Ts, Elongation factor Tu, ... | | 著者 | Gytz, H, Seweryn, P, Kutlubaeva, Z, Chetverin, A.B, Brodersen, D.E, Knudsen, C.R. | | 登録日 | 2014-08-26 | | 公開日 | 2015-09-23 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.21 Å) | | 主引用文献 | Structural basis for RNA-genome recognition during bacteriophage Q beta replication.
Nucleic Acids Res., 43, 2015
|
|
6VMY
 
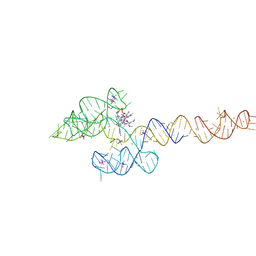 | | Structure of the B. subtilis cobalamin riboswitch | | 分子名称: | Adenosylcobalamin, B. subtilis cobalamin riboswitch, COBALT HEXAMMINE(III), ... | | 著者 | Chan, C.W, Mondragon, A. | | 登録日 | 2020-01-28 | | 公開日 | 2020-06-10 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | Crystal structure of an atypical cobalamin riboswitch reveals RNA structural adaptability as basis for promiscuous ligand binding.
Nucleic Acids Res., 48, 2020
|
|
3AX1
 
 | |
8T1R
 
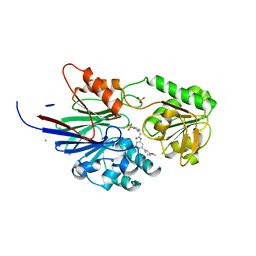 | | Crystal structure of human CPSF73 catalytic segment in complex with compound 2 | | 分子名称: | 3-[7,7-bis(oxidanyl)-8-oxa-7-boranuidabicyclo[4.3.0]nona-1,3,5-trien-5-yl]-~{N}-[3-(3-methoxyphenyl)phenyl]propanamide, CHLORIDE ION, Cleavage and polyadenylation specificity factor subunit 3, ... | | 著者 | Huang, J, Tong, L. | | 登録日 | 2023-06-02 | | 公開日 | 2023-11-08 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Anticancer benzoxaboroles block pre-mRNA processing by directly inhibiting CPSF3.
Cell Chem Biol, 31, 2024
|
|
8T1Q
 
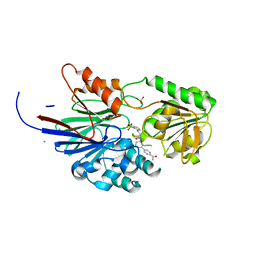 | | Crystal structure of human CPSF73 catalytic segment in complex with compound 1 | | 分子名称: | 3-[7,7-bis(oxidanyl)-8-oxa-7-boranuidabicyclo[4.3.0]nona-1,3,5-trien-5-yl]-~{N}-[3-(4-ethanoylphenyl)phenyl]propanamide, CHLORIDE ION, Cleavage and polyadenylation specificity factor subunit 3, ... | | 著者 | Huang, J, Tong, L. | | 登録日 | 2023-06-02 | | 公開日 | 2023-11-08 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Anticancer benzoxaboroles block pre-mRNA processing by directly inhibiting CPSF3.
Cell Chem Biol, 31, 2024
|
|
6Y83
 
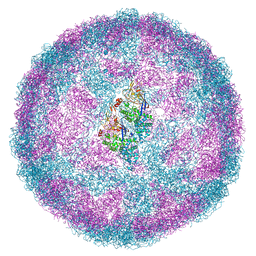 | | Capsid structure of Leishmania RNA virus 1 | | 分子名称: | Capsid protein | | 著者 | Prochazkova, M, Fuzik, T, Grybtchuk, D, Falginella, F, Podesvova, L, Yurchenko, V, Vacha, R, Plevka, P. | | 登録日 | 2020-03-03 | | 公開日 | 2020-11-11 | | 最終更新日 | 2022-03-30 | | 実験手法 | ELECTRON MICROSCOPY (3.65 Å) | | 主引用文献 | Capsid Structure of Leishmania RNA Virus 1.
J.Virol., 95, 2021
|
|
4KL5
 
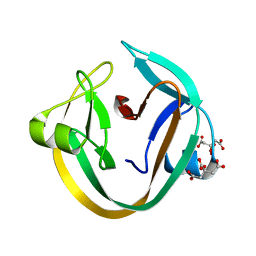 | | Crystal structure of NpuDnaE intein | | 分子名称: | CITRIC ACID, DNA polymerase III, alpha subunit, ... | | 著者 | Aranko, A.S, Oeemig, J.S, Kajander, T, Iwai, H. | | 登録日 | 2013-05-07 | | 公開日 | 2013-09-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Intermolecular domain swapping induces intein-mediated protein alternative splicing.
Nat.Chem.Biol., 9, 2013
|
|
4LCK
 
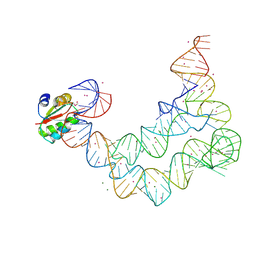 | |
7Z90
 
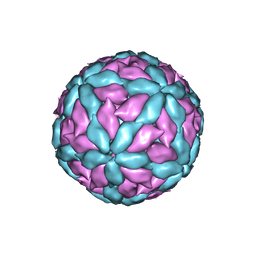 | |
7TNY
 
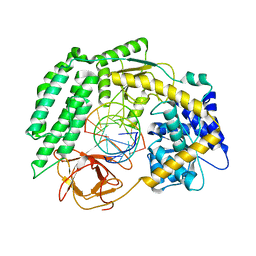 | | Cryo-EM structure of RIG-I in complex with p2dsRNA | | 分子名称: | Antiviral innate immune response receptor RIG-I, ZINC ION, p2dsRNA | | 著者 | Wang, W, Pyle, A.M. | | 登録日 | 2022-01-22 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
7TNX
 
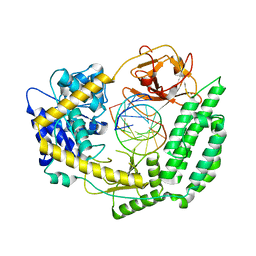 | | Cryo-EM structure of RIG-I in complex with p3dsRNA | | 分子名称: | Antiviral innate immune response receptor RIG-I, ZINC ION, p3dsRNAa, ... | | 著者 | Wang, W, Pyle, A.M. | | 登録日 | 2022-01-22 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.54 Å) | | 主引用文献 | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
7TO0
 
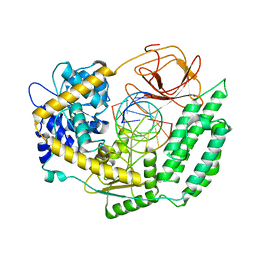 | | Cryo-EM structure of RIG-I in complex with OHdsRNA | | 分子名称: | Antiviral innate immune response receptor RIG-I, OHdsRNA, ZINC ION | | 著者 | Wang, W, Pyle, A.M. | | 登録日 | 2022-01-22 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
7TNZ
 
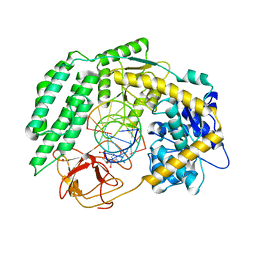 | | Cryo-EM structure of RIG-I in complex with p1dsRNA | | 分子名称: | Antiviral innate immune response receptor RIG-I, ZINC ION, p1dsRNA | | 著者 | Wang, W, Pyle, A.M. | | 登録日 | 2022-01-22 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.54 Å) | | 主引用文献 | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
2KTZ
 
 | | Inhibitor Induced Structural Change in the HCV IRES Domain IIa RNA | | 分子名称: | (7R)-7-[(dimethylamino)methyl]-1-[3-(dimethylamino)propyl]-7,8-dihydro-1H-furo[3,2-e]benzimidazol-2-amine, HCV IRES Domain IIa RNA | | 著者 | Paulsen, R.B, Seth, P.P, Swayze, E.E, Griffey, R.H, Skalicky, J.J, Cheatham III, T.E, Davis, D.R. | | 登録日 | 2010-02-10 | | 公開日 | 2010-04-28 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Inhibitor-induced structural change in the HCV IRES domain IIa RNA.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2KU0
 
 | | Inhibitor Induced Structural Change in the HCV IRES Domain IIa RNA | | 分子名称: | (7S)-7-[(dimethylamino)methyl]-1-[3-(dimethylamino)propyl]-7,8-dihydro-1H-furo[3,2-e]benzimidazol-2-amine, HCV IRES Domain IIa RNA | | 著者 | Paulsen, R.B, Seth, P.P, Swayze, E.E, Griffey, R.H, Skalicky, J.J, Cheatham III, T.E, Davis, D.R. | | 登録日 | 2010-02-10 | | 公開日 | 2010-04-28 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Inhibitor-induced structural change in the HCV IRES domain IIa RNA.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6D3P
 
 | | Crystal structure of an exoribonuclease-resistant RNA from Sweet clover necrotic mosaic virus (SCNMV) | | 分子名称: | IRIDIUM HEXAMMINE ION, RNA (45-MER) | | 著者 | Steckelberg, A.-L, Akiyama, B.M, Costantino, D.A, Sit, T.L, Nix, J.C, Kieft, J.S. | | 登録日 | 2018-04-16 | | 公開日 | 2018-06-20 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | A folded viral noncoding RNA blocks host cell exoribonucleases through a conformationally dynamic RNA structure.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3NPN
 
 | |
