4NU6
 
 | | Crystal Structure of PTDH R301K | | 分子名称: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Phosphonate dehydrogenase, SULFATE ION | | 著者 | Nair, S.K, Chekan, J.R. | | 登録日 | 2013-12-03 | | 公開日 | 2014-03-12 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Chemical rescue and inhibition studies to determine the role of arg301 in phosphite dehydrogenase.
Plos One, 9, 2014
|
|
4F2G
 
 | | The Crystal Structure of Ornithine carbamoyltransferase from Burkholderia thailandensis E264 | | 分子名称: | 1,2-ETHANEDIOL, Ornithine carbamoyltransferase 1, PHOSPHATE ION | | 著者 | Craig, T.K, Fox, D, Staker, B, Stewart, L, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2012-05-07 | | 公開日 | 2012-05-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Combining functional and structural genomics to sample the essential Burkholderia structome.
Plos One, 8, 2013
|
|
4NWR
 
 | | Computationally Designed Two-Component Self-Assembling Tetrahedral Cage T33-28 | | 分子名称: | Macrophage migration inhibitory factor-like protein, integron gene cassette protein | | 著者 | McNamara, D.E, King, N.P, Bale, J.B, Sheffler, W, Baker, D, Yeates, T.O. | | 登録日 | 2013-12-06 | | 公開日 | 2014-05-28 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Accurate design of co-assembling multi-component protein nanomaterials.
Nature, 510, 2014
|
|
4NVI
 
 | |
4NWO
 
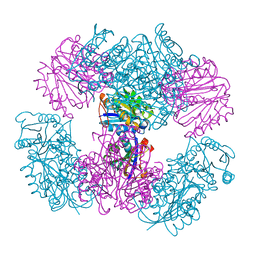 | | Computationally Designed Two-Component Self-Assembling Tetrahedral Cage T33-15 | | 分子名称: | CALCIUM ION, Chorismate mutase AroH, Molybdenum cofactor biosynthesis protein MogA | | 著者 | McNamara, D.E, King, N.P, Bale, J.B, Sheffler, W, Baker, D, Yeates, T.O. | | 登録日 | 2013-12-06 | | 公開日 | 2014-05-28 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Accurate design of co-assembling multi-component protein nanomaterials.
Nature, 510, 2014
|
|
4F47
 
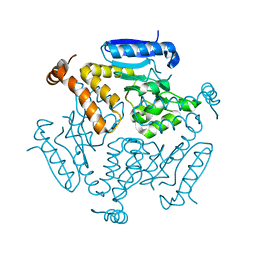 | |
4O45
 
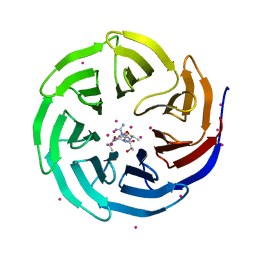 | | WDR5 in complex with influenza NS1 C-terminal tail | | 分子名称: | Nonstructural protein 1, UNKNOWN ATOM OR ION, WD repeat-containing protein 5 | | 著者 | Qin, S, Xu, C, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2013-12-18 | | 公開日 | 2014-04-23 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Structural basis for histone mimicry and hijacking of host proteins by influenza virus protein NS1.
Nat Commun, 5, 2014
|
|
4O64
 
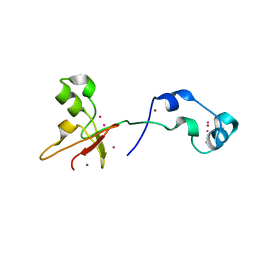 | | Zinc fingers of KDM2B | | 分子名称: | Lysine-specific demethylase 2B, UNKNOWN ATOM OR ION, ZINC ION | | 著者 | Liu, K, Xu, C, Tempel, W, Walker, J.R, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2013-12-20 | | 公開日 | 2014-04-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | DNA Sequence Recognition of Human CXXC Domains and Their Structural Determinants.
Structure, 26, 2018
|
|
4F49
 
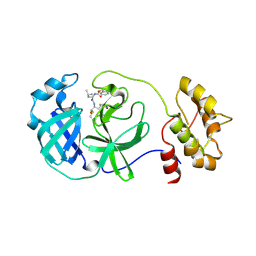 | | 2.25A resolution structure of Transmissible Gastroenteritis Virus Protease containing a covalently bound Dipeptidyl Inhibitor | | 分子名称: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, TETRAETHYLENE GLYCOL | | 著者 | Lovell, S, Battaile, K.P, Kim, Y, Tiew, K.-C, Mandadapu, S.R, Alliston, K.R, Groutas, W.C, Chang, K.-O. | | 登録日 | 2012-05-10 | | 公開日 | 2012-09-05 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Broad-Spectrum Antivirals against 3C or 3C-Like Proteases of Picornaviruses, Noroviruses, and Coronaviruses.
J.Virol., 86, 2012
|
|
4F76
 
 | |
4NTI
 
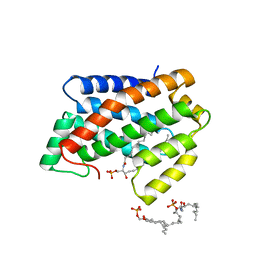 | | Crystal structure of D60N mutant of Arabidopsis ACD11 (accelerated-cell-death 11) complexed with C12 ceramide-1-phosphate (d18:1/12:0) at 2.9 Angstrom resolution | | 分子名称: | (2S,3R,4E)-2-(dodecanoylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, DI(HYDROXYETHYL)ETHER, accelerated-cell-death 11 | | 著者 | Simanshu, D.K, Brown, R.E, Patel, D.J. | | 登録日 | 2013-12-02 | | 公開日 | 2014-02-05 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.899 Å) | | 主引用文献 | Arabidopsis Accelerated Cell Death 11, ACD11, Is a Ceramide-1-Phosphate Transfer Protein and Intermediary Regulator of Phytoceramide Levels.
Cell Rep, 6, 2014
|
|
4F7V
 
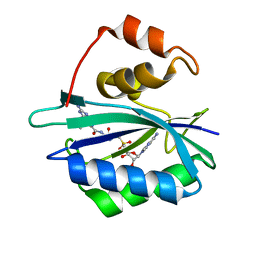 | | Crystal structure of E. coli HPPK in complex with bisubstrate analogue inhibitor J1D (HP26) | | 分子名称: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 5'-{[2-({N-[(2-amino-7,7-dimethyl-4-oxo-3,4,7,8-tetrahydropteridin-6-yl)carbonyl]glycyl}amino)ethyl]sulfonyl}-5'-deoxyadenosine | | 著者 | Shaw, G, Shi, G, Ji, X. | | 登録日 | 2012-05-16 | | 公開日 | 2012-07-11 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Bisubstrate analog inhibitors of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase: New lead exhibits a distinct binding mode.
Bioorg.Med.Chem., 20, 2012
|
|
4NWC
 
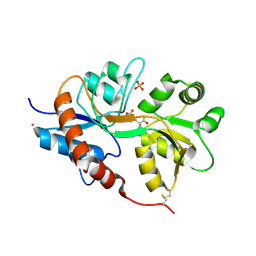 | | Crystal structure of the GluK3 ligand-binding domain (S1S2) in complex with the agonist (2S,4R)-4-(3-Methoxy-3-oxopropyl)glutamic acid at 2.01 A resolution. | | 分子名称: | (2S,4R)-4-(3-Methoxy-3-oxopropyl) glutamic acid, CHLORIDE ION, Glutamate receptor ionotropic, ... | | 著者 | Larsen, A.P, Venskutonyte, R, Frydenvang, K, Gajhede, M, Kastrup, J.S. | | 登録日 | 2013-12-06 | | 公開日 | 2014-08-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.012 Å) | | 主引用文献 | Molecular Recognition of Two 2,4-syn-Functionalized (S)-Glutamate Analogues by the Kainate Receptor GluK3 Ligand Binding Domain.
Chemmedchem, 9, 2014
|
|
4O5X
 
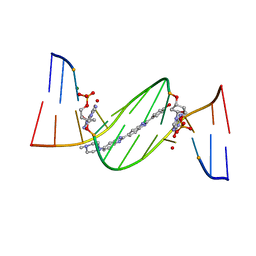 | | O6-carboxymethylguanine in DNA forms a sequence context dependent wobble base pair structure with thymine. | | 分子名称: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*(C6G)P*AP*AP*TP*TP*TP*GP*CP*G)-3'), MAGNESIUM ION | | 著者 | Zhang, F, Tsunoda, M, Suzuki, K, Kikuchi, Y, Wilkinson, O, Millington, C.L, Margison, G.P, Williams, D.M, Takenaka, A. | | 登録日 | 2013-12-20 | | 公開日 | 2014-07-02 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | O(6)-Carboxymethylguanine in DNA forms a sequence context-dependent wobble base-pair structure with thymine
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4F36
 
 | |
4F3X
 
 | | Crystal structure of putative aldehyde dehydrogenase from Sinorhizobium meliloti 1021 complexed with NAD | | 分子名称: | GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative aldehyde dehydrogenase | | 著者 | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2012-05-09 | | 公開日 | 2012-05-30 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Crystal structure of putative aldehyde dehydrogenase from Sinorhizobium meliloti 1021 complexed with NAD
To be Published
|
|
4FFW
 
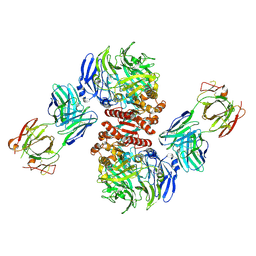 | | Crystal Structure of Dipeptidyl Peptidase IV (DPP4, DPP-IV, CD26) in Complex with Fab + sitagliptin | | 分子名称: | (2R)-4-OXO-4-[3-(TRIFLUOROMETHYL)-5,6-DIHYDRO[1,2,4]TRIAZOLO[4,3-A]PYRAZIN-7(8H)-YL]-1-(2,4,5-TRIFLUOROPHENYL)BUTAN-2-A MINE, Dipeptidyl peptidase 4, Fab heavy chain, ... | | 著者 | Wang, Z, Sudom, A, Walker, N.P, Min, X. | | 登録日 | 2012-06-01 | | 公開日 | 2012-12-12 | | 最終更新日 | 2021-05-19 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | An Inhibitory Antibody Against DPP IV Improves Glucose Tolerance in vivo - Validation of Large Molecule Approach for DPP IV Inhibition
To be published
|
|
4F82
 
 | |
3LN7
 
 | |
3LH1
 
 | |
3LV0
 
 | |
3L6D
 
 | | Crystal structure of putative oxidoreductase from Pseudomonas putida KT2440 | | 分子名称: | Putative oxidoreductase | | 著者 | Malashkevich, V.N, Patskovsky, Y, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-12-23 | | 公開日 | 2010-01-12 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of putative oxidoreductase from Pseudomonas putida KT2440
To be Published
|
|
3L6Y
 
 | | Crystal structure of p120 catenin in complex with E-cadherin | | 分子名称: | Catenin delta-1, E-cadherin | | 著者 | Ishiyama, N, Lee, S.-H, Liu, S, Li, G.-Y, Smith, M.J, Reichardt, L.F, Ikura, M. | | 登録日 | 2009-12-27 | | 公開日 | 2010-04-21 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Dynamic and static interactions between p120 catenin and E-cadherin regulate the stability of cell-cell adhesion.
Cell(Cambridge,Mass.), 141, 2010
|
|
3LX5
 
 | | Crystal structure of mGMPPNP-bound NFeoB from S. thermophilus | | 分子名称: | 2-amino-9-(5-O-[(R)-hydroxy{[(R)-hydroxy(phosphonoamino)phosphoryl]oxy}phosphoryl]-3-O-{[2-(methylamino)phenyl]carbonyl}-beta-D-erythro-pentofuranosyl-2-ulose)-1,9-dihydro-6H-purin-6-one, Ferrous iron uptake transporter protein B, GLYCEROL, ... | | 著者 | Ash, M.R, Guilfoyle, A, Maher, M.J, Clarke, R.J, Guss, J.M, Jormakka, M. | | 登録日 | 2010-02-24 | | 公開日 | 2010-03-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Potassium-activated GTPase reaction in the G Protein-coupled ferrous iron transporter B.
J.Biol.Chem., 285, 2010
|
|
3L81
 
 | | Crystal structure of adaptor protein complex 4 (AP-4) mu4 subunit C-terminal domain, in complex with a sorting peptide from the amyloid precursor protein (APP) | | 分子名称: | AP-4 complex subunit mu-1, Amyloid beta A4 protein, GLYCEROL | | 著者 | Mardones, G.A, Rojas, A.L, Burgos, P.V, Dasilva, L.L.P, Prabhu, Y, Bonifacino, J.S, Hurley, J.H. | | 登録日 | 2009-12-29 | | 公開日 | 2010-06-02 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Sorting of the Alzheimer's disease amyloid precursor protein mediated by the AP-4 complex.
Dev.Cell, 18, 2010
|
|
