2G10
 
 | | Photolyzed CO L29F Myoglobin: 3.16ns | | 分子名称: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Aranda, R, Levin, E.J, Schotte, F, Anfinrud, P.A, Phillips Jr, G.N. | | 登録日 | 2006-02-13 | | 公開日 | 2006-07-04 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Time-dependent atomic coordinates for the dissociation of carbon monoxide from myoglobin.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2GG6
 
 | | CP4 EPSP synthase liganded with S3P | | 分子名称: | 3-phosphoshikimate 1-carboxyvinyltransferase, SHIKIMATE-3-PHOSPHATE, SULFATE ION | | 著者 | Schonbrunn, E, Funke, T. | | 登録日 | 2006-03-23 | | 公開日 | 2006-08-22 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Molecular basis for the herbicide resistance of Roundup Ready crops.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
5KNK
 
 | | Lipid A secondary acyltransferase LpxM from Acinetobacter baumannii with catalytic residue substitution (E127A) | | 分子名称: | DODECYL-BETA-D-MALTOSIDE, GLYCEROL, Lipid A biosynthesis lauroyl acyltransferase, ... | | 著者 | Dovala, D.L, Hu, Q, Metzger IV, L.E. | | 登録日 | 2016-06-28 | | 公開日 | 2016-09-28 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure-guided enzymology of the lipid A acyltransferase LpxM reveals a dual activity mechanism.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6DDW
 
 | | Mycobacterium tuberculosis Dihydrofolate Reductase complexed with beta-NADPH and N-(4-{[(2-amino-4-oxo-3,4-dihydropteridin-6-yl)methyl]amino}-2-hydroxybenzene-1-carbonyl)-L-glutamic acid | | 分子名称: | Dihydrofolate reductase, GLYCEROL, N-(4-{[(2-amino-4-oxo-3,4-dihydropteridin-6-yl)methyl]amino}-2-hydroxybenzene-1-carbonyl)-L-glutamic acid, ... | | 著者 | Hajian, B, Scocchera, E, Wright, D. | | 登録日 | 2018-05-10 | | 公開日 | 2018-05-23 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Drugging the Folate Pathway in Mycobacterium tuberculosis: The Role of Multi-targeting Agents.
Cell Chem Biol, 26, 2019
|
|
5L5Q
 
 | |
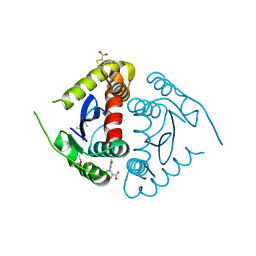
5KRT
 
 | |
2G7Q
 
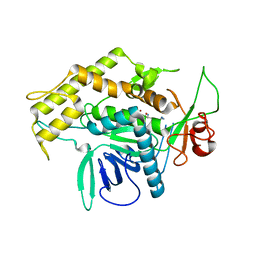 | | Structure of the Light Chain of Botulinum Neurotoxin Serotype A Bound to Small Molecule Inhibitors | | 分子名称: | Botulinum neurotoxin type A, N-HYDROXY-L-ARGININAMIDE, ZINC ION | | 著者 | Fu, Z, Baldwin, M.R, Boldt, G.E, Janda, K.D, Barbieri, J.T, Kim, J.-J.P. | | 登録日 | 2006-02-28 | | 公開日 | 2006-08-15 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Light chain of botulinum neurotoxin serotype A: structural resolution of a catalytic intermediate.
Biochemistry, 45, 2006
|
|
5L66
 
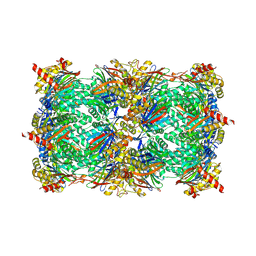 | |
2G5K
 
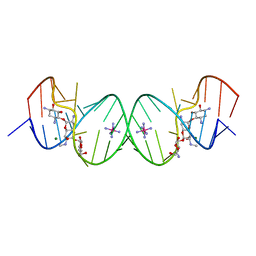 | | Crystal Structure of the Homo sapiens Cytoplasmic Ribosomal Decoding Site complexed with Apramycin | | 分子名称: | 5'-R(*UP*UP*GP*CP*GP*UP*CP*GP*CP*UP*CP*CP*GP*GP*AP*AP*AP*AP*GP*UP*CP*GP*C)-3', APRAMYCIN, COBALT HEXAMMINE(III), ... | | 著者 | Kondo, J, Francois, B, Urzhumtsev, A, Westhof, E. | | 登録日 | 2006-02-23 | | 公開日 | 2006-06-20 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal Structure of the Homo sapiens Cytoplasmic Ribosomal Decoding Site Complexed with Apramycin
Angew.Chem.Int.Ed.Engl., 45, 2006
|
|
2G9X
 
 | |
2G5Z
 
 | | Structure of S65G Y66S GFP variant after spontaneous peptide hydrolysis and decarboxylation | | 分子名称: | Green fluorescent protein, MAGNESIUM ION | | 著者 | Barondeau, D.P, Kassmann, C.J, Tainer, J.A, Getzoff, E.D. | | 登録日 | 2006-02-23 | | 公開日 | 2006-04-18 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Understanding GFP Posttranslational Chemistry: Structures of Designed Variants that Achieve Backbone Fragmentation, Hydrolysis, and Decarboxylation.
J.Am.Chem.Soc., 128, 2006
|
|
5L5O
 
 | | Yeast 20S proteasome with human beta5i (1-138) and human beta6 (97-111; 118-133) in complex with epoxyketone inhibitor 16 | | 分子名称: | (2~{S})-3-(1~{H}-indol-3-yl)-~{N}-[(2~{S},3~{S},4~{R})-4-methyl-3,5-bis(oxidanyl)-1-phenyl-pentan-2-yl]-2-[[(2~{R})-2-(2-morpholin-4-ylethanoylamino)propanoyl]amino]propanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | 著者 | Groll, M, Huber, E.M. | | 登録日 | 2016-05-28 | | 公開日 | 2016-11-09 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | A humanized yeast proteasome identifies unique binding modes of inhibitors for the immunosubunit beta 5i.
EMBO J., 35, 2016
|
|
5L65
 
 | |
2GHW
 
 | | Crystal structure of SARS spike protein receptor binding domain in complex with a neutralizing antibody, 80R | | 分子名称: | CHLORIDE ION, Spike glycoprotein, anti-sars scFv antibody, ... | | 著者 | Hwang, W.C, Lin, Y, Santelli, E, Sui, J, Jaroszewski, L, Stec, B, Farzan, M, Marasco, W.A, Liddington, R.C. | | 登録日 | 2006-03-27 | | 公開日 | 2006-09-19 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis of neutralization by a human anti-severe acute respiratory syndrome spike protein antibody, 80R.
J.Biol.Chem., 281, 2006
|
|
2G9P
 
 | | NMR structure of a novel antimicrobial peptide, latarcin 2a, from spider (Lachesana tarabaevi) venom | | 分子名称: | antimicrobial peptide Latarcin 2a | | 著者 | Dubovskii, P.V, Volynsky, P.E, Polyansky, A.A, Chupin, V.V, Efremov, R.G, Arseniev, A.S. | | 登録日 | 2006-03-07 | | 公開日 | 2006-09-12 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Spatial structure and activity mechanism of a novel spider antimicrobial peptide.
Biochemistry, 45, 2006
|
|
6LHY
 
 | | Crystal structure of ThsB | | 分子名称: | DUF1863 domain-containing protein | | 著者 | Bae, E, Ka, D, Oh, H. | | 登録日 | 2019-12-10 | | 公開日 | 2020-06-24 | | 実験手法 | X-RAY DIFFRACTION (1.796 Å) | | 主引用文献 | Structural and functional evidence of bacterial antiphage protection by Thoeris defense system via NAD+degradation.
Nat Commun, 11, 2020
|
|
4UMT
 
 | | Structure of MELK in complex with inhibitors | | 分子名称: | 1-(4-{[3-(isoquinolin-7-yl)prop-2-yn-1-yl]oxy}-2-methoxybenzyl)piperazinediium, DIMETHYL SULFOXIDE, MATERNAL EMBRYONIC LEUCINE ZIPPER KINASE | | 著者 | Johnson, C.N, Berdini, V, Beke, L, Bonnet, P, Brehmer, D, Coyle, J.E, Day, P.J, Frederickson, M, Freyne, E.J.E, Gilissen, R.A.H.J, Hamlett, C.C.F, Howard, S, Meerpoel, L, McMenamin, R, Patel, S, Rees, D.C, Sharff, A, Sommen, F, Wu, T, Linders, J.T.M. | | 登録日 | 2014-05-21 | | 公開日 | 2014-10-22 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Structure-Based Design of Type II Inhibitors Applied to Maternal Embryonic Leucine Zipper Kinase.
Acs Med.Chem.Lett., 6, 2015
|
|
4UMU
 
 | | Structure of MELK in complex with inhibitors | | 分子名称: | (2-ethoxy-4-{[3-(isoquinolin-7-yl)prop-2-yn-1-yl]oxy}phenyl)methanaminium, MATERNAL EMBRYONIC LEUCINE ZIPPER KINASE | | 著者 | Johnson, C.N, Berdini, V, Beke, L, Bonnet, P, Brehmer, D, Coyle, J.E, Day, P.J, Frederickson, M, Freyne, E.J.E, Gilissen, R.A.H.J, Hamlett, C.C.F, Howard, S, Meerpoel, L, McMenamin, R, Patel, S, Rees, D.C, Sharff, A, Sommen, F, Wu, T, Linders, J.T.M. | | 登録日 | 2014-05-21 | | 公開日 | 2014-10-22 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Structure-Based Design of Type II Inhibitors Applied to Maternal Embryonic Leucine Zipper Kinase.
Acs Med.Chem.Lett., 6, 2015
|
|
4UMP
 
 | | Structure of MELK in complex with inhibitors | | 分子名称: | 3-(isoquinolin-7-yl)prop-2-yn-1-ol, MATERNAL EMBRYONIC LEUCINE ZIPPER KINASE | | 著者 | Johnson, C.N, Berdini, V, Beke, L, Bonnet, P, Brehmer, D, Coyle, J.E, Day, P.J, Frederickson, M, Freyne, E.J.E, Gilissen, R.A.H.J, Hamlett, C.C.F, Howard, S, Meerpoel, L, McMenamin, R, Patel, S, Rees, D.C, Sharff, A, Sommen, F, Wu, T, Linders, J.T.M. | | 登録日 | 2014-05-20 | | 公開日 | 2014-10-08 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Fragment-Based Discovery of Type I Inhibitors of Maternal Embryonic Leucine Zipper Kinase
Acs Med.Chem.Lett., 6, 2015
|
|
4USE
 
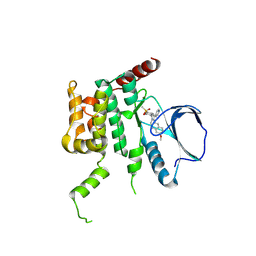 | | Human STK10 (LOK) with SB-633825 | | 分子名称: | 4-{5-(6-methoxynaphthalen-2-yl)-1-methyl-2-[2-methyl-4-(methylsulfonyl)phenyl]-1H-imidazol-4-yl}pyridine, SERINE/THREONINE-PROTEIN KINASE 10 | | 著者 | Elkins, J.M, Salah, E, Szklarz, M, von Delft, F, Canning, P, Raynor, J, Bountra, C, Edwards, A.M, Knapp, S. | | 登録日 | 2014-07-07 | | 公開日 | 2015-07-22 | | 最終更新日 | 2018-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Comprehensive Characterization of the Published Kinase Inhibitor Set.
Nat.Biotechnol., 34, 2016
|
|
4UUK
 
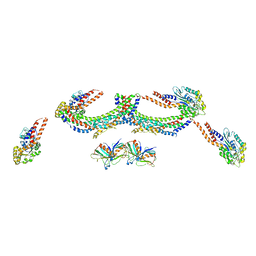 | | Human dynamin 1 K44A superconstricted polymer stabilized with GTP strand 2 | | 分子名称: | DYNAMIN-1 | | 著者 | Sundborger, A.C, Fang, S, Heymann, J.A, Ray, P, Chappie, J.S, Hinshaw, J.E. | | 登録日 | 2014-07-29 | | 公開日 | 2014-08-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (12.5 Å) | | 主引用文献 | A Dynamin Mutant Defines a Superconstricted Prefission State.
Cell Rep., 8, 2014
|
|
4V0G
 
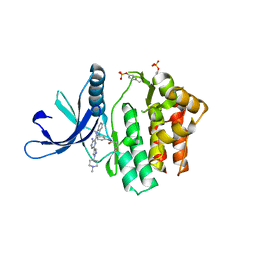 | | JAK3 in complex with a covalent EGFR inhibitor | | 分子名称: | N-[3-(2-{3-amino-6-[1-(1-methylpiperidin-4-yl)-1H-pyrazol-4-yl]pyrazin-2-yl}-1H-benzimidazol-1-yl)phenyl]propanamide, TYROSINE-PROTEIN KINASE JAK3 | | 著者 | Debreczeni, J.E, Hennessy, E.J, Chuaquini, C, Ashton, S, Coclough, N, Cross, D.A.E, Eberlein, C, Gingipalli, L, Klinowska, T.C.M, Orme, J.P, Sha, L, Wu, X. | | 登録日 | 2014-09-16 | | 公開日 | 2016-01-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Utilisation of Structure Based Design to Identify Novel, Irreversible Inhibitors of the Epidermal Growth Factor Receptor (Egfr) Harboring the Gatekeeper T790M Mutation
To be Published
|
|
7XOJ
 
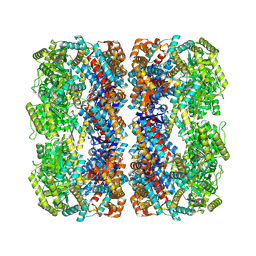 | |
7XOK
 
 | |
7X3Y
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle in complex with nAb 9A3 (CVB1-E:9A3) | | 分子名称: | 9A3 heavy chain, 9A3 light chain, VP2, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-03-01 | | 公開日 | 2023-06-07 | | 実験手法 | ELECTRON MICROSCOPY (3.32 Å) | | 主引用文献 | Cryo-EM structure of Coxsackievirus B1 empty particle in complex with nAb 9A3 (CVB1-E:9A3)
To Be Published
|
|
