3TX0
 
 | | Unphosphorylated Bacillus cereus phosphopentomutase in a P212121 crystal form | | 分子名称: | MANGANESE (II) ION, Phosphopentomutase | | 著者 | Panosian, T.P, Nanneman, D.P, Bachmann, B.O, Iverson, T.M. | | 登録日 | 2011-09-22 | | 公開日 | 2012-02-29 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Molecular Differences between a Mutase and a Phosphatase: Investigations of the Activation Step in Bacillus cereus Phosphopentomutase.
Biochemistry, 51, 2012
|
|
2MS8
 
 | |
1N3G
 
 | |
4BQ0
 
 | | Pseudomonas aeruginosa beta-alanine:pyruvate aminotransferase holoenzyme without divalent cations on dimer-dimer interface | | 分子名称: | BETA-ALANINE--PYRUVATE TRANSAMINASE, CHLORIDE ION, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Isupov, M.N, Lebedev, A.A, Westlake, A, Sayer, C, Littlechild, J.A. | | 登録日 | 2013-05-29 | | 公開日 | 2013-06-05 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Space-Group and Origin Ambiguity in Macromolecular Structures with Pseudo-Symmetry and its Treatment with the Program Zanuda.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4GRT
 
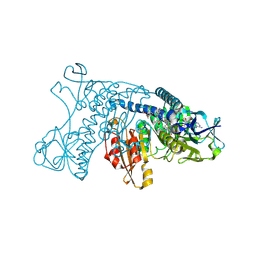 | | HUMAN GLUTATHIONE REDUCTASE A34E, R37W MUTANT, MIXED DISULFIDE BETWEEN TRYPANOTHIONE AND THE ENZYME | | 分子名称: | BIS(GAMMA-GLUTAMYL-CYSTEINYL-GLYCINYL)SPERMIDINE, FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE REDUCTASE | | 著者 | Stoll, V.S, Simpson, S.J, Krauth-Siegel, R.L, Walsh, C.T, Pai, E.F. | | 登録日 | 1997-02-12 | | 公開日 | 1997-08-12 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Glutathione reductase turned into trypanothione reductase: structural analysis of an engineered change in substrate specificity.
Biochemistry, 36, 1997
|
|
1PDF
 
 | | Fitting of gp11 crystal structure into 3D cryo-EM reconstruction of bacteriophage T4 baseplate-tail tube complex | | 分子名称: | Baseplate structural protein Gp11 | | 著者 | Kostyuchenko, V.A, Leiman, P.G, Chipman, P.R, Kanamaru, S, van Raaij, M.J, Arisaka, F, Mesyanzhinov, V.V, Rossmann, M.G. | | 登録日 | 2003-05-19 | | 公開日 | 2003-09-09 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (12 Å) | | 主引用文献 | Three-dimensional structure of bacteriophage T4 baseplate
Nat.Struct.Biol., 10, 2003
|
|
1P7J
 
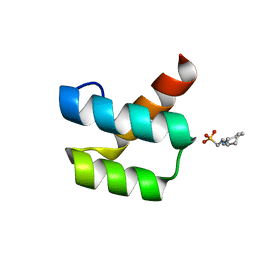 | | Crystal structure of engrailed homeodomain mutant K52E | | 分子名称: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Segmentation polarity homeobox protein engrailed | | 著者 | Stollar, E.J, Mayor, U, Lovell, S.C, Federici, L, Freund, S.M, Fersht, A.R, Luisi, B.F. | | 登録日 | 2003-05-02 | | 公開日 | 2003-10-14 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structures of Engrailed Homeodomain Mutants: IMPLICATIONS FOR STABILITY AND DYNAMICS
J.Biol.Chem., 278, 2003
|
|
4HVP
 
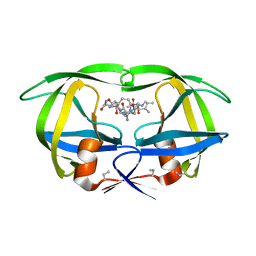 | | Structure of complex of synthetic HIV-1 protease with a substrate-based inhibitor at 2.3 Angstroms resolution | | 分子名称: | HIV-1 PROTEASE, N-{(2S)-2-[(N-acetyl-L-threonyl-L-isoleucyl)amino]hexyl}-L-norleucyl-L-glutaminyl-N~5~-[amino(iminio)methyl]-L-ornithinamide | | 著者 | Miller, M, Schneider, J, Sathyanarayana, B.K, Toth, M.V, Marshall, G.R, Clawson, L, Selk, L, Kent, S.B.H, Wlodawer, A. | | 登録日 | 1989-08-08 | | 公開日 | 1990-04-15 | | 最終更新日 | 2017-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of complex of synthetic HIV-1 protease with a substrate-based inhibitor at 2.3 A resolution.
Science, 246, 1989
|
|
4ID1
 
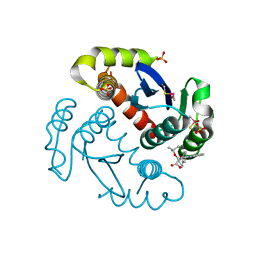 | |
2BO5
 
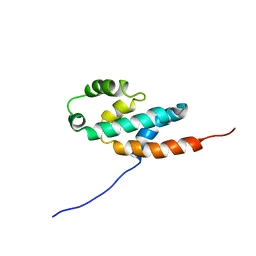 | | Bovine oligomycin sensitivity conferral protein N-terminal domain | | 分子名称: | ATP SYNTHASE OLIGOMYCIN SENSITIVITY CONFERRAL PROTEIN | | 著者 | Carbajo, R.J, Kellas, F.A, Runswick, M.J, Montgomery, M.G, Walker, J.E, Neuhaus, D. | | 登録日 | 2005-04-07 | | 公開日 | 2005-08-17 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the F1-binding domain of the stator of bovine F1Fo-ATPase and how it binds an alpha-subunit.
J. Mol. Biol., 351, 2005
|
|
2HF6
 
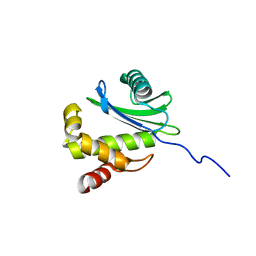 | | Solution structure of human zeta-COP | | 分子名称: | Coatomer subunit zeta-1 | | 著者 | Yu, W, Jin, C, Xia, B. | | 登録日 | 2006-06-23 | | 公開日 | 2007-06-12 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of human zeta-COP: direct evidences for structural similarity between COP I and clathrin-adaptor coats
J.Mol.Biol., 386, 2009
|
|
4C0B
 
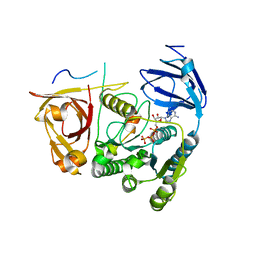 | |
1P7I
 
 | | CRYSTAL STRUCTURE OF ENGRAILED HOMEODOMAIN MUTANT K52A | | 分子名称: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Segmentation polarity homeobox protein engrailed | | 著者 | Stollar, E.J, Mayor, U, Lovell, S.C, Federici, L, Freund, S.M, Fersht, A.R, Luisi, B.F. | | 登録日 | 2003-05-02 | | 公開日 | 2003-10-14 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structures of Engrailed Homeodomain Mutants: IMPLICATIONS FOR STABILITY AND DYNAMICS
J.Biol.Chem., 278, 2003
|
|
1PYD
 
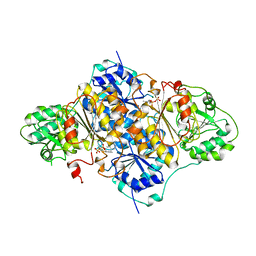 | |
4CT0
 
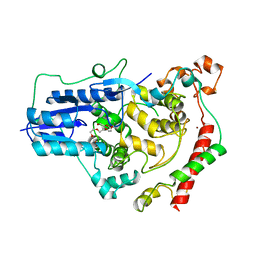 | | Crystal Structure of Mouse Cryptochrome1 in Complex with Period2 | | 分子名称: | CHLORIDE ION, CRYPTOCHROME-1, HEXAETHYLENE GLYCOL, ... | | 著者 | Schmalen, I, Rajan Prabu, J, Benda, C, Wolf, E. | | 登録日 | 2014-03-11 | | 公開日 | 2014-06-04 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Interaction of Circadian Clock Proteins Cry1 and Per2 is Modulated by Zinc Binding and Disulfide Bond Formation.
Cell(Cambridge,Mass.), 157, 2014
|
|
3GRT
 
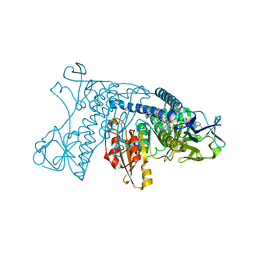 | | HUMAN GLUTATHIONE REDUCTASE A34E, R37W MUTANT, OXIDIZED TRYPANOTHIONE COMPLEX | | 分子名称: | 2-AMINO-4-[4-(4-AMINO-4-CARBOXY-BUTYRYLAMINO)-5,8,19,22-TETRAOXO-1,2-DITHIA-6,9,13,18,21-PENTAAZA-CYCLOTETRACOS-23-YLCARBAMOYL]-BUTYRIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE REDUCTASE | | 著者 | Stoll, V.S, Simpson, S.J, Krauth-Siegel, R.L, Walsh, C.T, Pai, E.F. | | 登録日 | 1997-02-12 | | 公開日 | 1997-08-12 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Glutathione reductase turned into trypanothione reductase: structural analysis of an engineered change in substrate specificity.
Biochemistry, 36, 1997
|
|
2LTS
 
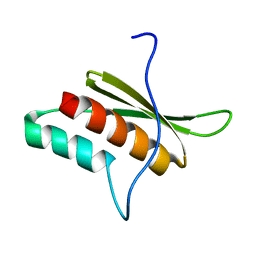 | |
2LTR
 
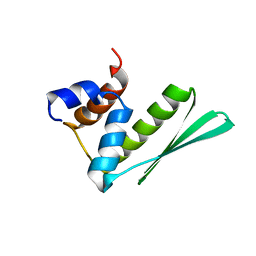 | | Solution structure of RDE-4(32-136) | | 分子名称: | Protein RDE-4 | | 著者 | Deshmukh, M, Chiliveri, S. | | 登録日 | 2012-05-31 | | 公開日 | 2013-12-04 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of RDE-4 dsRBDs and mutational studies provide insights into dsRNA recognition in the Caenorhabditis elegans RNAi pathway.
Biochem.J., 458, 2014
|
|
387D
 
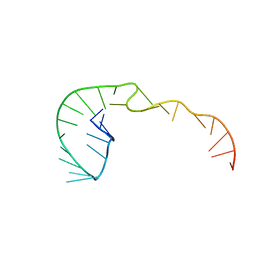 | | RNA Pseudoknot with 3D Domain Swapping | | 分子名称: | RNA Pseudoknot | | 著者 | Lietzke, S.E, Kundrot, C.E, Barnes, C.L. | | 登録日 | 1998-04-14 | | 公開日 | 2003-08-26 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | The Structure of an RNA Pseudoknot Shows 3D Domain Swapping
Structure, Motion, Interaction and Expression of Biological Macromolecules, The Proceedings of the Tenth Conversation held at The University-SUNY, Albany NY, June 17-21, 1997, 10, 1998
|
|
3A99
 
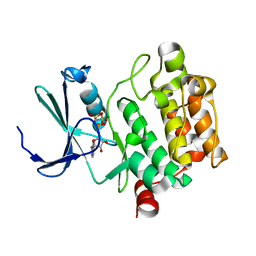 | | Structure of PIM-1 kinase crystallized in the presence of P27KIP1 Carboxy-terminal peptide | | 分子名称: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Proto-oncogene serine/threonine-protein kinase pim-1 | | 著者 | Morishita, D, Takami, M, Yoshikawa, S, Katayama, R, Sato, S, Kukimoto-Niino, M, Umehara, T, Shirouzu, M, Sekimizu, K, Yokoyama, S, Fujita, N. | | 登録日 | 2009-10-22 | | 公開日 | 2010-11-03 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Cell-permeable carboxyl-terminal p27(Kip1) peptide exhibits anti-tumor activity by inhibiting Pim-1 kinase
J.Biol.Chem., 286, 2011
|
|
3B3X
 
 | | Crystal structure of class A beta-lactamase of Bacillus licheniformis BS3 with aminocitrate | | 分子名称: | 2-(carboxymethyl)-D-aspartic acid, Beta-lactamase | | 著者 | Sauvage, E, Herman, R, Kerff, F, Charlier, P. | | 登録日 | 2007-10-23 | | 公開日 | 2007-11-13 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | 2-Aminopropane-1,2,3-tricarboxylic acid: Synthesis and co-crystallization with the class A beta-lactamase BS3 of Bacillus licheniformis
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3BGP
 
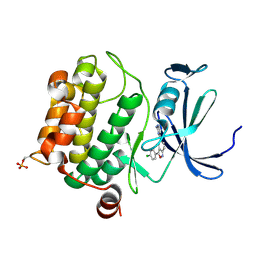 | | Human Pim-1 complexed with a benzoisoxazole inhibitor VX1 | | 分子名称: | 4-[3-(4-chlorophenyl)-2,1-benzisoxazol-5-yl]pyrimidin-2-amine, Proto-oncogene serine/threonine-protein kinase Pim-1 | | 著者 | Jacobs, M.D. | | 登録日 | 2007-11-27 | | 公開日 | 2007-12-11 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Docking study yields four novel inhibitors of the protooncogene pim-1 kinase.
J.Med.Chem., 51, 2008
|
|
3IUH
 
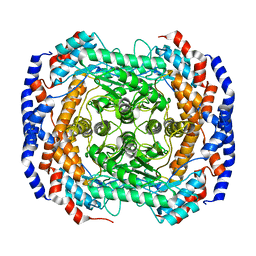 | | Co2+-bound form of Pseudomonas stutzeri L-rhamnose isomerase | | 分子名称: | COBALT (II) ION, L-rhamnose isomerase | | 著者 | Yoshida, H, Yamaji, M, Ishii, T, Izumori, K, Kamitori, S. | | 登録日 | 2009-08-31 | | 公開日 | 2010-02-02 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Catalytic reaction mechanism of Pseudomonas stutzeri l-rhamnose isomerase deduced from X-ray structures
Febs J., 277, 2010
|
|
3IYZ
 
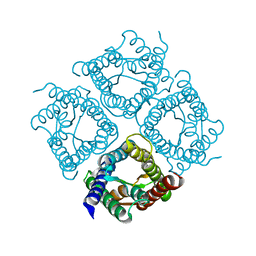 | | Structure of Aquaporin-4 S180D mutant at 10.0 A resolution from electron micrograph | | 分子名称: | Aquaporin-4 | | 著者 | Mitsuma, T, Tani, K, Hiroaki, Y, Kamegawa, A, Suzuki, H, Hibino, H, Kurachi, Y, Fujiyoshi, Y. | | 登録日 | 2010-07-24 | | 公開日 | 2010-08-25 | | 最終更新日 | 2023-09-06 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (10 Å) | | 主引用文献 | Influence of the cytoplasmic domains of aquaporin-4 on water conduction and array formation.
J.Mol.Biol., 402, 2010
|
|
3J97
 
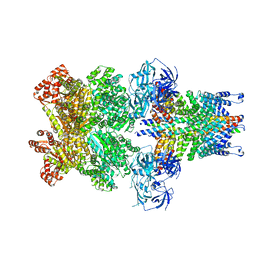 | | Structure of 20S supercomplex determined by single particle cryoelectron microscopy (State II) | | 分子名称: | Alpha-soluble NSF attachment protein, Synaptosomal-associated protein 25, Syntaxin-1A, ... | | 著者 | Zhao, M, Wu, S, Cheng, Y, Brunger, A.T. | | 登録日 | 2014-12-05 | | 公開日 | 2015-01-28 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (7.8 Å) | | 主引用文献 | Mechanistic insights into the recycling machine of the SNARE complex.
Nature, 518, 2015
|
|
