1DCU
 
 | | REDOX SIGNALING IN THE CHLOROPLAST: STRUCTURE OF OXIDIZED PEA FRUCTOSE-1,6-BISPHOSPHATE PHOSPHATASE | | 分子名称: | FRUCTOSE-1,6-BISPHOSPHATASE | | 著者 | Chiadmi, M, Navaza, A, Miginiac-Maslow, M, Jacquot, J.P, Cherfils, J. | | 登録日 | 1999-11-05 | | 公開日 | 1999-12-03 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Redox signalling in the chloroplast: structure of oxidized pea fructose-1,6-bisphosphate phosphatase.
EMBO J., 18, 1999
|
|
2GCI
 
 | | The 1,1-proton transfer reaction mechanism by alpha-methylacyl-CoA racemase is catalyzed by an asparte/histidine pair and involves a smooth, methionine-rich surface for binding the fatty acyl moiety | | 分子名称: | (R)-2-METHYLMYRISTOYL-COENZYME A, GLYCEROL, probable alpha-methylacyl-CoA racemase MCR | | 著者 | Bhaumik, P, Wierenga, R.K. | | 登録日 | 2006-03-14 | | 公開日 | 2007-02-20 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The Catalysis of the 1,1-Proton Transfer by alpha-Methyl-acyl-CoA Racemase Is Coupled to a Movement of the Fatty Acyl Moiety Over a Hydrophobic, Methionine-rich Surface
J.Mol.Biol., 367, 2007
|
|
1WJA
 
 | | SOLUTION STRUCTURE OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE (D FORM), NMR, REGULARIZED MEAN STRUCTURE | | 分子名称: | HIV-1 INTEGRASE, ZINC ION | | 著者 | Clore, G.M, Cai, M, Caffrey, M, Gronenborn, A.M. | | 登録日 | 1997-05-13 | | 公開日 | 1998-05-13 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the N-terminal zinc binding domain of HIV-1 integrase.
Nat.Struct.Biol., 4, 1997
|
|
4CKR
 
 | | Crystal structure of the human DDR1 kinase domain in complex with DDR1-IN-1 | | 分子名称: | 1,2-ETHANEDIOL, 4-[(4-ethylpiperazin-1-yl)methyl]-n-{4-methyl-3-[(2-oxo-2,3-dihydro-1h-indol-5-yl)oxy]phenyl}-3-(trifluoromethyl)benzamide, EPITHELIAL DISCOIDIN DOMAIN-CONTAINING RECEPTOR 1 | | 著者 | Canning, P, Elkins, J.M, Goubin, S, Mahajan, P, Krojer, T, Newman, J.A, Dixon-Clarke, S, Chaikuad, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | 登録日 | 2014-01-07 | | 公開日 | 2014-01-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Discovery of a Potent and Selective Ddr1 Receptor Tyrosine Kinase Inhibitor.
Acs Chem.Biol., 8, 2013
|
|
3IV0
 
 | |
3WQJ
 
 | | Crystal structure of archaerhodopsin-2 at 1.8 angstrom resolution | | 分子名称: | (6E,10E,14E,18E)-2,6,10,15,19,23-hexamethyltetracosa-2,6,10,14,18,22-hexaene, 2,3-DI-O-PHYTANLY-3-SN-GLYCERO-1-PHOSPHORYL-3'-SN-GLYCEROL-1'-PHOSPHATE, 2,3-DI-PHYTANYL-GLYCEROL, ... | | 著者 | Kouyama, T. | | 登録日 | 2014-01-27 | | 公開日 | 2014-10-15 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of archaerhodopsin-2 at 1.8 angstrom resolution.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WVJ
 
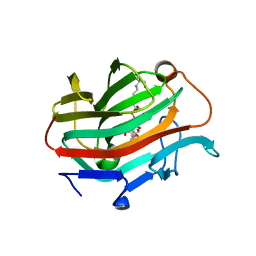 | | The crystal structure of native glycosidic hydrolase | | 分子名称: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucanase | | 著者 | Chen, C.C, Huang, J.W, Zhao, P, Ko, T.P, Huang, C.H, Chan, H.C, Huang, Z, Liu, W, Cheng, Y.S, Liu, J.R, Guo, R.T. | | 登録日 | 2014-05-22 | | 公開日 | 2015-06-24 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural analyses and yeast production of the beta-1,3-1,4-glucanase catalytic module encoded by the licB gene of Clostridium thermocellum.
Enzyme.Microb.Technol., 71, 2015
|
|
3EL1
 
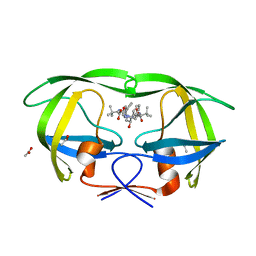 | | Crystal Structure of wild-type HIV protease in complex with the inhibitor, Atazanavir | | 分子名称: | (3S,8S,9S,12S)-3,12-BIS(1,1-DIMETHYLETHYL)-8-HYDROXY-4,11-DIOXO-9-(PHENYLMETHYL)-6-[[4-(2-PYRIDINYL)PHENYL]METHYL]-2,5, 6,10,13-PENTAAZATETRADECANEDIOIC ACID DIMETHYL ESTER, ACETATE ION, ... | | 著者 | Schiffer, C.A, Nalam, M.N.L. | | 登録日 | 2008-09-19 | | 公開日 | 2009-09-01 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Extreme Entropy-Enthalpy Compensation in a Drug-Resistant Variant of HIV-1 Protease.
Acs Chem.Biol., 7, 2012
|
|
3EKY
 
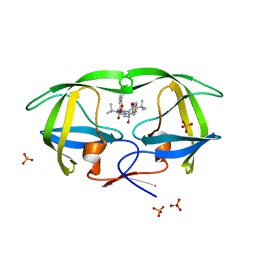 | | Crystal Structure of wild-type HIV protease in complex with the inhibitor, Atazanavir | | 分子名称: | (3S,8S,9S,12S)-3,12-BIS(1,1-DIMETHYLETHYL)-8-HYDROXY-4,11-DIOXO-9-(PHENYLMETHYL)-6-[[4-(2-PYRIDINYL)PHENYL]METHYL]-2,5, 6,10,13-PENTAAZATETRADECANEDIOIC ACID DIMETHYL ESTER, PHOSPHATE ION, ... | | 著者 | Schiffer, C.A, Nalam, M.N.L. | | 登録日 | 2008-09-19 | | 公開日 | 2009-09-01 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Extreme Entropy-Enthalpy Compensation in a Drug-Resistant Variant of HIV-1 Protease.
Acs Chem.Biol., 7, 2012
|
|
3BXR
 
 | | Crystal Structures Of Highly Constrained Substrate And Hydrolysis Products Bound To HIV-1 Protease. Implications For Catalytic Mechanism | | 分子名称: | (9S,12S)-9-(1-methylethyl)-N-[(8S,11S)-8-[(1S)-1-methylpropyl]-7,10-dioxo-2-oxa-6,9-diazabicyclo[11.2.2]heptadeca-1(15),13,16-trien-11-yl]-7,10-dioxo-2-oxa-8,11-diazabicyclo[12.2.2]octadeca-1(16),14,17-triene-12-carboxamide, Protease, SULFATE ION | | 著者 | Tyndall, J.D, Pattenden, L.K, Reid, R.C, Hu, S.H, Alewood, D, Alewood, P.F, Walsh, T, Fairlie, D.P, Martin, J.L. | | 登録日 | 2008-01-14 | | 公開日 | 2008-03-25 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal Structures of Highly Constrained Substrate and Hydrolysis Products Bound to HIV-1 Protease. Implications for the Catalytic Mechanism
Biochemistry, 47, 2008
|
|
10GS
 
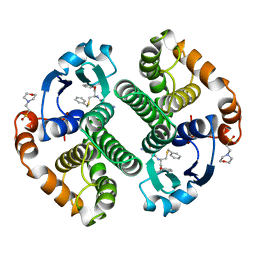 | | HUMAN GLUTATHIONE S-TRANSFERASE P1-1, COMPLEX WITH TER117 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE S-TRANSFERASE P1-1, L-gamma-glutamyl-S-benzyl-N-[(S)-carboxy(phenyl)methyl]-L-cysteinamide | | 著者 | Oakley, A, Parker, M. | | 登録日 | 1997-08-14 | | 公開日 | 1998-09-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The structures of human glutathione transferase P1-1 in complex with glutathione and various inhibitors at high resolution.
J.Mol.Biol., 274, 1997
|
|
19GS
 
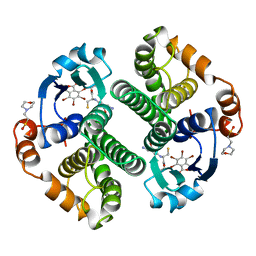 | | Glutathione s-transferase p1-1 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3,3'-(4,5,6,7-TETRABROMO-3-OXO-1(3H)-ISOBENZOFURANYLIDENE)BIS [6-HYDROXYBENZENESULFONIC ACID]ANION, GLUTATHIONE, ... | | 著者 | Oakley, A.J, Lo Bello, M, Parker, M.W. | | 登録日 | 1997-12-14 | | 公開日 | 1998-12-30 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The ligandin (non-substrate) binding site of human Pi class glutathione transferase is located in the electrophile binding site (H-site).
J.Mol.Biol., 291, 1999
|
|
3TIH
 
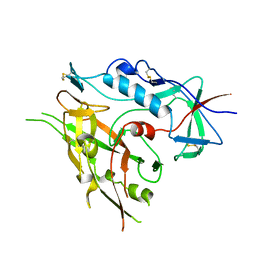 | |
3C5T
 
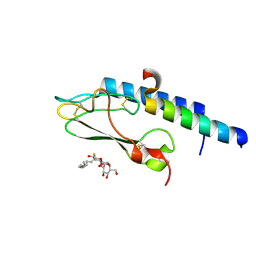 | |
1R0A
 
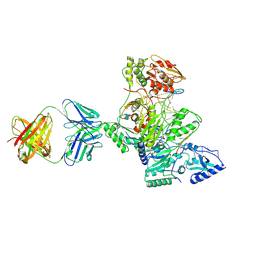 | | Crystal structure of HIV-1 reverse transcriptase covalently tethered to DNA template-primer solved to 2.8 angstroms | | 分子名称: | 5'-D(*A*TP*GP*CP*AP*TP*CP*GP*GP*CP*GP*CP*TP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*GP*GP*T)-3', 5'-D(*C*CP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*AP*GP*CP*GP*CP*CP*GP*(2DA))-3', GLYCEROL, ... | | 著者 | Tuske, S, Ding, J, Arnold, E. | | 登録日 | 2003-09-19 | | 公開日 | 2004-08-03 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Nonnucleoside inhibitor binding affects the interactions of the fingers subdomain of human immunodeficiency virus type 1 reverse transcriptase with DNA.
J.Virol., 78, 2004
|
|
1XYN
 
 | |
3UZC
 
 | | Thermostabilised Adenosine A2A receptor in complex with 4-(3-amino-5-phenyl-1,2,4-triazin-6-yl)-2-chlorophenol | | 分子名称: | 4-(3-amino-5-phenyl-1,2,4-triazin-6-yl)-2-chlorophenol, Adenosine A2A Receptor | | 著者 | Congreve, M, Andrews, S.P, Dore, A.S, Hollenstein, K, Hurrell, E, Langmead, C.J, Mason, J.S, Ng, I.W, Zhukov, A, Weir, M, Marshall, F.H. | | 登録日 | 2011-12-07 | | 公開日 | 2012-03-21 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (3.341 Å) | | 主引用文献 | Discovery of 1,2,4-Triazine Derivatives as Adenosine A(2A) Antagonists using Structure Based Drug Design
J.Med.Chem., 55, 2012
|
|
1BWN
 
 | | PH DOMAIN AND BTK MOTIF FROM BRUTON'S TYROSINE KINASE MUTANT E41K IN COMPLEX WITH INS(1,3,4,5)P4 | | 分子名称: | BRUTON'S TYROSINE KINASE, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, ZINC ION | | 著者 | Djinovic Carugo, K, Baraldi, E, Hyvoenen, M, Lo Surdo, P, Riley, A, Potter, B, Saraste, M. | | 登録日 | 1998-09-25 | | 公開日 | 1999-06-15 | | 最終更新日 | 2024-12-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of the PH domain from Bruton's tyrosine kinase in complex with inositol 1,3,4,5-tetrakisphosphate.
Structure Fold.Des., 7, 1999
|
|
2FYD
 
 | | catalytic domain of bovine beta 1, 4-galactosyltransferase in complex with alpha-lactalbumin, glucose, Mn, and UDP-N-acetylgalactosamine | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-galactopyranose, Alpha-lactalbumin, ... | | 著者 | Ramakrishnan, B, Ramasamy, V, Qasba, P.K. | | 登録日 | 2006-02-07 | | 公開日 | 2006-03-14 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Snapshots of beta-1,4-Galactosyltransferase-I Along the Kinetic Pathway.
J.Mol.Biol., 357, 2006
|
|
3UYT
 
 | | crystal structure of ck1d with PF670462 from P1 crystal form | | 分子名称: | 4-[1-cyclohexyl-4-(4-fluorophenyl)-1H-imidazol-5-yl]pyrimidin-2-amine, Casein kinase I isoform delta, SULFATE ION | | 著者 | Huang, X. | | 登録日 | 2011-12-06 | | 公開日 | 2012-01-11 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis for the interaction between casein kinase 1 delta and a potent and selective inhibitor.
J.Med.Chem., 55, 2012
|
|
4EWO
 
 | | Design and synthesis of potent hydroxyethylamine (hea) bace-1 inhibitors | | 分子名称: | Beta-secretase 1, N-[(2S,3R)-4-{[(4S)-2-(2,2-dimethylpropyl)-6,6-dimethyl-4,5,6,7-tetrahydro-2H-indazol-4-yl]amino}-3-hydroxy-1-phenylbutan-2-yl]acetamide | | 著者 | Borkakoti, N, Lindberg, J, Derbyshire, D. | | 登録日 | 2012-04-27 | | 公開日 | 2012-10-31 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Design and synthesis of potent hydroxyethylamine (HEA) BACE-1 inhibitors carrying prime side 4,5,6,7-tetrahydrobenzazole and 4,5,6,7-tetrahydropyridinoazole templates.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
7EKZ
 
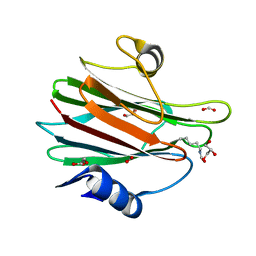 | | Structural and functional insights into Hydra Actinoporin-Like Toxin 1 (HALT-1) | | 分子名称: | FORMIC ACID, GLYCEROL, HALT-1 | | 著者 | Ker, D.S, Sha, X.H, Jonet, M.A, Hwang, J.S, Ng, C.L. | | 登録日 | 2021-04-07 | | 公開日 | 2022-02-16 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | Structural and functional analysis of Hydra Actinoporin-Like Toxin 1 (HALT-1).
Sci Rep, 11, 2021
|
|
3IUP
 
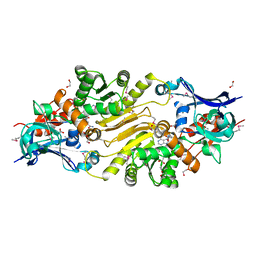 | |
3AL1
 
 | | DESIGNED PEPTIDE ALPHA-1, RACEMIC P1BAR FORM | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ETHANOLAMINE, PROTEIN (D, ... | | 著者 | Patterson, W.R, Anderson, D.H, Degrado, W.F, Cascio, D, Eisenberg, D. | | 登録日 | 1998-10-26 | | 公開日 | 1998-11-04 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (0.75 Å) | | 主引用文献 | Centrosymmetric bilayers in the 0.75 A resolution structure of a designed alpha-helical peptide, D,L-Alpha-1.
Protein Sci., 8, 1999
|
|
4EXG
 
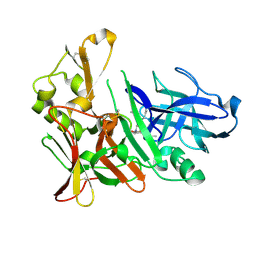 | | Design and synthesis of potent hydroxyethylamine (hea) bace-1 inhibitors | | 分子名称: | Beta-secretase 1, N-[(2S,3R)-4-{[(4S)-6-(2,2-dimethylpropyl)-2,2-dimethyl-3,4-dihydro-2H-thieno[2,3-b]pyran-4-yl]amino}-3-hydroxy-1-phenylbutan-2-yl]acetamide | | 著者 | Borkakoti, N, Lindberg, J, Derbyshire, D. | | 登録日 | 2012-04-30 | | 公開日 | 2012-10-31 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Design and synthesis of potent hydroxyethylamine (HEA) BACE-1 inhibitors carrying prime side 4,5,6,7-tetrahydrobenzazole and 4,5,6,7-tetrahydropyridinoazole templates.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
