4HPI
 
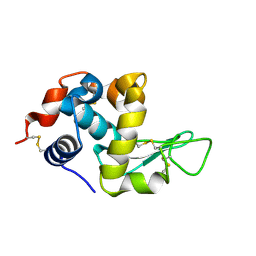 | | Crystal Structure of Hen Egg White Lysozyme complex with GN2-M | | 分子名称: | 1-DEOXYNOJIRIMYCIN, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysozyme C | | 著者 | Umemoto, N, Numata, T, Ohnuma, T, Fukamizo, T. | | 登録日 | 2012-10-23 | | 公開日 | 2013-01-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.19 Å) | | 主引用文献 | A novel transition-state analogue for lysozyme, 4-O-beta-tri-N-acetylchitotriosyl moranoline, provided evidence supporting the covalent glycosyl-enzyme intermediate.
J.Biol.Chem., 288, 2013
|
|
6TL3
 
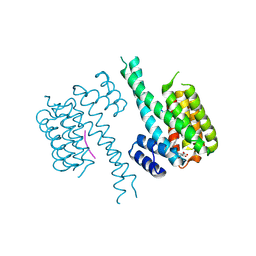 | | Crystal structure of an Estrogen Receptor alpha 8-mer phosphopeptide in complex with 14-3-3sigma stabilized by a Pyrrolidone1 derivative | | 分子名称: | 14-3-3 protein sigma, 5-[(2~{S},3~{R})-3-[(~{R})-azanyl(phenyl)methyl]-2-(4-nitrophenyl)-4,5-bis(oxidanylidene)pyrrolidin-1-yl]-2-oxidanyl-benzoic acid, Estrogen receptor | | 著者 | Andrei, S.A, Bosica, F, Ottmann, C, O'Mahony, G. | | 登録日 | 2019-11-30 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.455 Å) | | 主引用文献 | Design of Drug-Like Protein-Protein Interaction Stabilizers Guided By Chelation-Controlled Bioactive Conformation Stabilization.
Chemistry, 26, 2020
|
|
1I6E
 
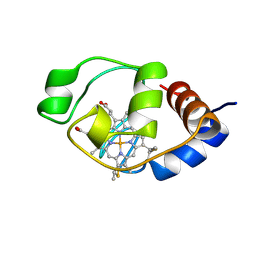 | | SOLUTION STRUCTURE OF THE FUNCTIONAL DOMAIN OF PARACOCCUS DENITRIFICANS CYTOCHROME C552 IN THE OXIDIZED STATE | | 分子名称: | CYTOCHROME C552, HEME C | | 著者 | Reincke, B, Perez, C, Pristovsek, P, Luecke, C, Ludwig, C, Loehr, F, Rogov, V.V, Ludwig, B, Rueterjans, H. | | 登録日 | 2001-03-02 | | 公開日 | 2001-10-17 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure and dynamics of the functional domain of Paracoccus denitrificans cytochrome c(552) in both redox states.
Biochemistry, 40, 2001
|
|
1DJ8
 
 | |
1KBQ
 
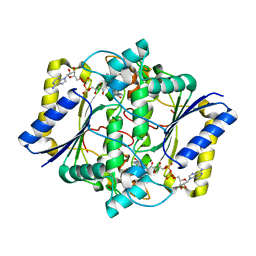 | | Complex of Human NAD(P)H quinone Oxidoreductase with 5-methoxy-1,2-dimethyl-3-(4-nitrophenoxymethyl)indole-4,7-dione (ES936) | | 分子名称: | 5-METHOXY-1,2-DIMETHYL-3-(4-NITROPHENOXYMETHYL)INDOLE-4,7-DIONE, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1 | | 著者 | Faig, M, Bianchet, M.A, Amzel, L.M. | | 登録日 | 2001-11-06 | | 公開日 | 2002-01-16 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Characterization of a mechanism-based inhibitor of NAD(P)H:quinone oxidoreductase 1 by biochemical, X-ray crystallographic, and mass spectrometric approaches.
Biochemistry, 40, 2001
|
|
1YHN
 
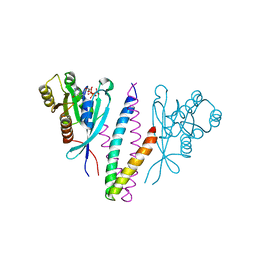 | | Structure basis of RILP recruitment by Rab7 | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Rab interacting lysosomal protein, ... | | 著者 | Wu, M, Wang, T, Hong, W, Song, H. | | 登録日 | 2005-01-10 | | 公開日 | 2005-05-17 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural basis for recruitment of RILP by small GTPase Rab7.
Embo J., 24, 2005
|
|
1RPK
 
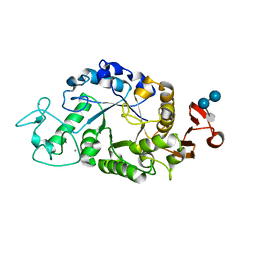 | | Crystal structure of barley alpha-amylase isozyme 1 (amy1) in complex with acarbose | | 分子名称: | 4,6-dideoxy-4-{[(1S,5R,6S)-3-formyl-5,6-dihydroxy-4-oxocyclohex-2-en-1-yl]amino}-alpha-D-xylo-hex-5-enopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-4-{[(1S,5R,6S)-3-formyl-5,6-dihydroxy-4-oxocyclohex-2-en-1-yl]amino}-alpha-D-xylo-hex-5-enopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-amylase type 1 isozyme, ... | | 著者 | Robert, X, Haser, R, Aghajari, N. | | 登録日 | 2003-12-03 | | 公開日 | 2005-06-07 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Oligosaccharide Binding to Barley {alpha}-Amylase 1
J.Biol.Chem., 280, 2005
|
|
1KBO
 
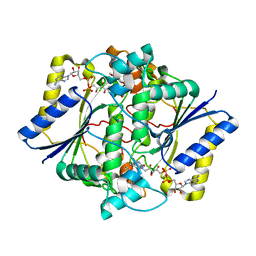 | | Complex of Human recombinant NAD(P)H:Quinone Oxide reductase type 1 with 5-methoxy-1,2-dimethyl-3-(phenoxymethyl)indole-4,7-dione (ES1340) | | 分子名称: | 5-METHOXY-1,2-DIMETHYL-3-(PHENOXYMETHYL)INDOLE-4,7-DIONE, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1 | | 著者 | Faig, M, Bianchet, M.A, Amzel, L.M. | | 登録日 | 2001-11-06 | | 公開日 | 2002-01-16 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Characterization of a mechanism-based inhibitor of NAD(P)H:quinone oxidoreductase 1 by biochemical, X-ray crystallographic, and mass spectrometric approaches.
Biochemistry, 40, 2001
|
|
4G32
 
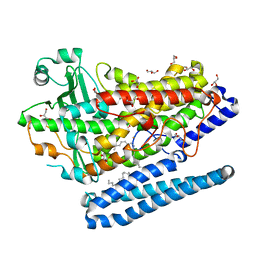 | | Crystal Structure of a Phospholipid-Lipoxygenase Complex from Pseudomonas aeruginosa at 1.75A (P21212) | | 分子名称: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradec-5-enoyloxy)propyl (11Z)-octadec-11-enoate, 15S-LIPOXYGENASE, FE (II) ION, ... | | 著者 | Carpena, X, Garreta, A, Val-Moraes, S.P, Garcia-Fernandez, Q, Fita, I. | | 登録日 | 2012-07-13 | | 公開日 | 2013-11-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structure and interaction with phospholipids of a prokaryotic lipoxygenase from Pseudomonas aeruginosa.
Faseb J., 27, 2013
|
|
4RIQ
 
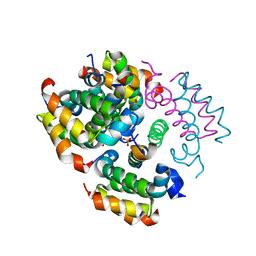 | |
1GM7
 
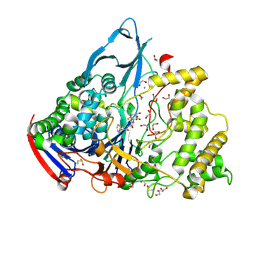 | | Crystal structures of penicillin acylase enzyme-substrate complexes: Structural insights into the catalytic mechanism | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, PENICILLIN G, ... | | 著者 | McVey, C.E, Walsh, M.A, Dodson, G.G, Wilson, K.S, Brannigan, J.A. | | 登録日 | 2001-09-11 | | 公開日 | 2001-11-28 | | 最終更新日 | 2019-07-24 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Crystal Structures of Penicillin Acylase Enzyme- Substrate Complexes: Structural Insights Into the Catalytic Mechanism
J.Mol.Biol., 313, 2001
|
|
4ITB
 
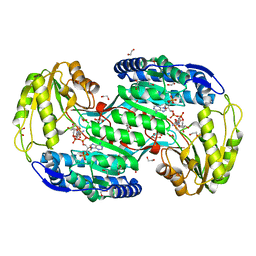 | | Structure of bacterial enzyme in complex with cofactor and substrate | | 分子名称: | 1,2-ETHANEDIOL, 4-oxobutanoic acid, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Rhee, S, Park, J. | | 登録日 | 2013-01-18 | | 公開日 | 2013-04-24 | | 最終更新日 | 2013-06-26 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural Basis for a Cofactor-dependent Oxidation Protection and Catalysis of Cyanobacterial Succinic Semialdehyde Dehydrogenase.
J.Biol.Chem., 288, 2013
|
|
1Z9E
 
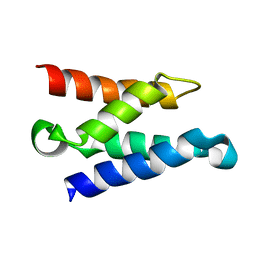 | | Solution structure of the HIV-1 integrase-binding domain in LEDGF/p75 | | 分子名称: | PC4 and SFRS1 interacting protein 2 | | 著者 | Cherepanov, P, Sun, Z.-Y.J, Rahman, S, Maertens, G, Wagner, G, Engelman, A. | | 登録日 | 2005-04-01 | | 公開日 | 2005-05-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the HIV-1 integrase-binding domain in LEDGF/p75
Nat.Struct.Mol.Biol., 12, 2005
|
|
4IY5
 
 | | Crystal structure of the glua2 ligand-binding domain (S1S2J-L483Y-N754S) in complex with glutamate and CX516 at 2.0 A resolution | | 分子名称: | CHLORIDE ION, GLUTAMIC ACID, GLYCEROL, ... | | 著者 | Krintel, C, Frydenvang, K, Harpsoe, K, Gajhede, M, Kastrup, J.S. | | 登録日 | 2013-01-28 | | 公開日 | 2013-10-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural analysis of the positive AMPA receptor modulators CX516 and Me-CX516 in complex with the GluA2 ligand-binding domain
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4IY6
 
 | | Crystal structure of the GLUA2 ligand-binding domain (S1S2J-L483Y-N754S) in complex with glutamate and ME-CX516 at 1.72 A resolution | | 分子名称: | GLUTAMIC ACID, GLYCEROL, Glutamate receptor 2, ... | | 著者 | Krintel, C, Frydenvang, K, Harpsoe, K, Gajhede, M, Kastrup, J.S. | | 登録日 | 2013-01-28 | | 公開日 | 2013-10-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Structural analysis of the positive AMPA receptor modulators CX516 and Me-CX516 in complex with the GluA2 ligand-binding domain
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4IT9
 
 | | Structure of Bacterial Enzyme | | 分子名称: | 1,2-ETHANEDIOL, PHOSPHATE ION, Succinate-semialdehyde dehydrogenase | | 著者 | Rhee, S, Park, J. | | 登録日 | 2013-01-18 | | 公開日 | 2013-04-24 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural Basis for a Cofactor-dependent Oxidation Protection and Catalysis of Cyanobacterial Succinic Semialdehyde Dehydrogenase.
J.Biol.Chem., 288, 2013
|
|
4ITA
 
 | | Structure of bacterial enzyme in complex with cofactor | | 分子名称: | 1,2-ETHANEDIOL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Succinate-semialdehyde dehydrogenase | | 著者 | Rhee, S, Park, J. | | 登録日 | 2013-01-18 | | 公開日 | 2013-04-24 | | 最終更新日 | 2013-06-26 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural Basis for a Cofactor-dependent Oxidation Protection and Catalysis of Cyanobacterial Succinic Semialdehyde Dehydrogenase.
J.Biol.Chem., 288, 2013
|
|
4IW3
 
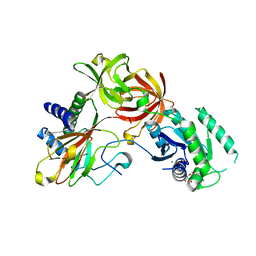 | | Crystal structure of a Pseudomonas putida prolyl-4-hydroxylase (P4H) in complex with elongation factor Tu (EF-Tu) | | 分子名称: | Elongation factor Tu-A, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Scotti, J.S, McDonough, M.A, Schofield, C.J. | | 登録日 | 2013-01-23 | | 公開日 | 2014-01-29 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.697 Å) | | 主引用文献 | Human oxygen sensing may have origins in prokaryotic elongation factor Tu prolyl-hydroxylation
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1VRU
 
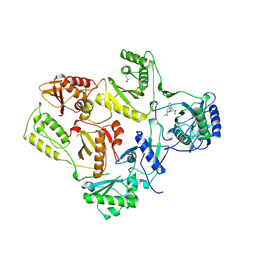 | | HIGH RESOLUTION STRUCTURES OF HIV-1 RT FROM FOUR RT-INHIBITOR COMPLEXES | | 分子名称: | ALPHA-(2,6-DICHLOROPHENYL)-ALPHA-(2-ACETYL-5-METHYLANILINO)ACETAMIDE, HIV-1 REVERSE TRANSCRIPTASE | | 著者 | Ren, J, Esnouf, R, Garman, E, Somers, D, Ross, C, Kirby, I, Keeling, J, Darby, G, Jones, Y, Stuart, D, Stammers, D. | | 登録日 | 1995-04-19 | | 公開日 | 1996-04-03 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | High resolution structures of HIV-1 RT from four RT-inhibitor complexes.
Nat.Struct.Biol., 2, 1995
|
|
1VRT
 
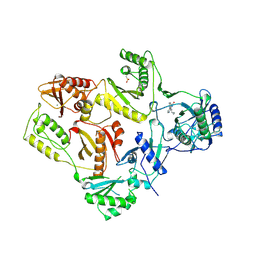 | | HIGH RESOLUTION STRUCTURES OF HIV-1 RT FROM FOUR RT-INHIBITOR COMPLEXES | | 分子名称: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE, MAGNESIUM ION | | 著者 | Ren, J, Esnouf, R, Garman, E, Somers, D, Ross, C, Kirby, I, Keeling, J, Darby, G, Jones, Y, Stuart, D, Stammers, D. | | 登録日 | 1995-04-19 | | 公開日 | 1996-04-03 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | High resolution structures of HIV-1 RT from four RT-inhibitor complexes.
Nat.Struct.Biol., 2, 1995
|
|
1LWE
 
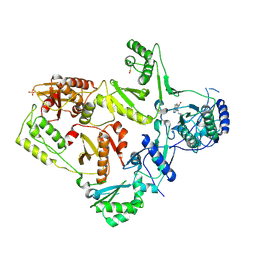 | | CRYSTAL STRUCTURE OF M41L/T215Y MUTANT HIV-1 REVERSE TRANSCRIPTASE (RTMN) IN COMPLEX WITH NEVIRAPINE | | 分子名称: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE, PHOSPHATE ION | | 著者 | Ren, J, Chamberlain, P.P, Nichols, C.E, Douglas, L, Stuart, D.I, Stammers, D.K. | | 登録日 | 2002-05-31 | | 公開日 | 2002-10-30 | | 最終更新日 | 2021-11-10 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | Crystal structures of Zidovudine- or Lamivudine-resistant human immunodeficiency virus type 1 reverse transcriptases containing mutations at codons 41, 184, and 215.
J.Virol., 76, 2002
|
|
4CAY
 
 | | Crystal structure of a human Anp32e-H2A.Z-H2B complex | | 分子名称: | ACIDIC LEUCINE-RICH NUCLEAR PHOSPHOPROTEIN 32 FAMILY MEMBER E, HISTONE H2A.Z, HISTONE H2B TYPE 1-J | | 著者 | Obri, A, Ouararhni, K, Papin, C, Diebold, M.-L, Padmanabhan, K, Marek, M, Stoll, I, Roy, L, Reilly, P.T, Mak, T.W, Dimitrov, S, Romier, C, Hamiche, A. | | 登録日 | 2013-10-09 | | 公開日 | 2014-01-22 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Anp32E is a Histone Chaperone that Removes H2A.Z from Chromatin
Nature, 648, 2014
|
|
3H4S
 
 | |
1L5A
 
 | | Crystal Structure of VibH, an NRPS Condensation Enzyme | | 分子名称: | amide synthase | | 著者 | Keating, T.A, Marshall, C.G, Walsh, C.T, Keating, A.E. | | 登録日 | 2002-03-06 | | 公開日 | 2002-06-26 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | The structure of VibH represents nonribosomal peptide synthetase condensation, cyclization and epimerization domains.
Nat.Struct.Biol., 9, 2002
|
|
7VH9
 
 | | Solution structure of the chimeric peptide of the first SURP domain of Human SF3A1 and the interacting region of SF1. | | 分子名称: | Splicing factor 3A subunit 1,Splicing factor 1 | | 著者 | Muto, Y, Kuwasako, K, Takizawa, M, Kobayashi, N, Sakamoto, T. | | 登録日 | 2021-09-21 | | 公開日 | 2022-09-28 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for the interaction between the first SURP domain of the SF3A1 subunit in U2 snRNP and the human splicing factor SF1.
Protein Sci., 31, 2022
|
|
