1VLR
 
 | |
1REE
 
 | | ENDO-1,4-BETA-XYLANASE II COMPLEX WITH 3,4-EPOXYBUTYL-BETA-D-XYLOSIDE | | 分子名称: | (3S)-3-hydroxybutyl beta-D-xylopyranoside, BENZOIC ACID, ENDO-1,4-BETA-XYLANASE II | | 著者 | Rouvinen, J, Havukainen, R, Torronen, A. | | 登録日 | 1995-12-21 | | 公開日 | 1997-01-11 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Covalent binding of three epoxyalkyl xylosides to the active site of endo-1,4-xylanase II from Trichoderma reesei.
Biochemistry, 35, 1996
|
|
3UFN
 
 | | Crystal Structure of Multidrug Resistant HIV-1 Protease Clinical Isolate PR20 in Complex with Saquinavir | | 分子名称: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, CHLORIDE ION, HIV-1 protease | | 著者 | Agniswamy, J, Chen-Hsiang, S, Aniana, A, Sayer, J.M, Louis, J.M, Weber, I.T. | | 登録日 | 2011-11-01 | | 公開日 | 2012-03-28 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | HIV-1 protease with 20 mutations exhibits extreme resistance to clinical inhibitors through coordinated structural rearrangements.
Biochemistry, 51, 2012
|
|
3RSV
 
 | |
1EUB
 
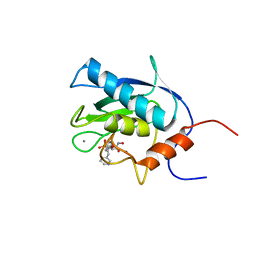 | | SOLUTION STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN COLLAGENASE-3 (MMP-13) COMPLEXED TO A POTENT NON-PEPTIDIC SULFONAMIDE INHIBITOR | | 分子名称: | 1-METHYLOXY-4-SULFONE-BENZENE, 3-METHYLPYRIDINE, CALCIUM ION, ... | | 著者 | Zhang, X, Gonnella, N.C, Koehn, J, Pathak, N, Ganu, V, Melton, R, Parker, D, Hu, S.I, Nam, K.Y. | | 登録日 | 2000-04-14 | | 公開日 | 2001-04-14 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the catalytic domain of human collagenase-3 (MMP-13) complexed to a potent non-peptidic sulfonamide inhibitor: binding comparison with stromelysin-1 and collagenase-1.
J.Mol.Biol., 301, 2000
|
|
3VBX
 
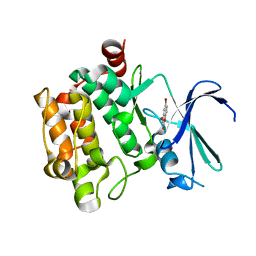 | |
3RTN
 
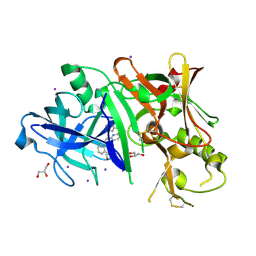 | |
3VBT
 
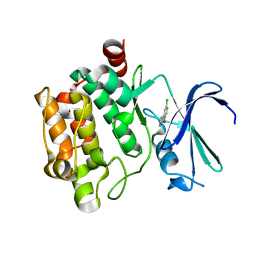 | |
3RJV
 
 | |
2A98
 
 | | Crystal structure of the catalytic domain of human inositol 1,4,5-trisphosphate 3-kinase C | | 分子名称: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate 3-kinase C | | 著者 | Hallberg, B.M, Ogg, D, Ehn, M, Graslund, S, Hammarstrom, M, Kotenyova, T, Nilsson-Ehle, P, Nordlund, P, Persson, C, Sagemark, J, Schuler, H, Stenmark, P, Thorsell, A.-G, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J, Structural Genomics Consortium (SGC) | | 登録日 | 2005-07-11 | | 公開日 | 2005-07-19 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The crystal structure of the catalytic domain of human inositol 1,4,5-trisphosphate 3-kinase C
To be Published
|
|
1VQZ
 
 | |
4K8K
 
 | | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with 1-(4-methoxyphenyl)-1-cyclopropane and 2-aminoperimidine | | 分子名称: | 1-(4-methoxyphenyl)cyclopropanecarboxylic acid, 1H-perimidin-2-amine, ADENOSINE, ... | | 著者 | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2013-04-18 | | 公開日 | 2013-05-01 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with 1-(4-methoxyphenyl)-1-cyclopropane and 2-aminoperimidine
To be Published
|
|
1EWE
 
 | |
2GFH
 
 | |
4OPX
 
 | | Structure of Human PARP-1 bound to a DNA double strand break in complex with (2R)-5-fluoro-2-methyl-2,3-dihydro-1-benzofuran-7-carboxamide | | 分子名称: | (2R)-5-fluoro-2-methyl-2,3-dihydro-1-benzofuran-7-carboxamide, DNA (26-MER), Poly [ADP-ribose] polymerase 1, ... | | 著者 | Pascal, J.M, Steffen, J.D. | | 登録日 | 2014-02-06 | | 公開日 | 2014-07-02 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.314 Å) | | 主引用文献 | Discovery and Structure-Activity Relationship of Novel 2,3-Dihydrobenzofuran-7-carboxamide and 2,3-Dihydrobenzofuran-3(2H)-one-7-carboxamide Derivatives as Poly(ADP-ribose)polymerase-1 Inhibitors.
J.Med.Chem., 57, 2014
|
|
3WIY
 
 | | Crystal structure of Mcl-1 in complex with compound 10 | | 分子名称: | 7-(4-{[(4-{[(2R)-4-(dimethylamino)-1-(phenylsulfanyl)butan-2-yl]amino}-3-nitrophenyl)sulfonyl]carbamoyl}-2-methylphenyl)-3-[3-(naphthalen-1-yloxy)propyl]pyrazolo[1,5-a]pyridine-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | 著者 | Sogabe, S, Igaki, S, Hayano, Y. | | 登録日 | 2013-09-26 | | 公開日 | 2013-11-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Discovery of potent Mcl-1/Bcl-xL dual inhibitors by using a hybridization strategy based on structural analysis of target proteins.
J.Med.Chem., 56, 2013
|
|
1NV5
 
 | | Fructose-1,6-Bisphosphatase Complex with Magnesium, Fructose-6-Phosphate, Phosphate, EDTA and Thallium (5 mM) | | 分子名称: | 6-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase, MAGNESIUM ION, ... | | 著者 | Choe, J, Iancu, C.V, Fromm, H.J, Honzatko, R.B. | | 登録日 | 2003-02-02 | | 公開日 | 2003-07-08 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Interaction of Tl+ with product complexes of fructose-1,6-bisphosphatase
J.BIOL.CHEM., 278, 2003
|
|
2G8N
 
 | | Structure of hPNMT with inhibitor 3-Hydroxymethyl-7-(N-4-chlorophenylaminosulfonyl)-THIQ and AdoHcy | | 分子名称: | (3R)-N-(4-CHLOROPHENYL)-3-(HYDROXYMETHYL)-1,2,3,4-TETRAHYDROISOQUINOLINE-7-SULFONAMIDE, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Drinkwater, N, Gee, C.L, Martin, J.L. | | 登録日 | 2006-03-02 | | 公開日 | 2006-09-12 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Comparison of the Binding of 3-Fluoromethyl-7-sulfonyl-1,2,3,4-tetrahydroisoquinolines with Their Isosteric Sulfonamides to the Active Site of Phenylethanolamine N-Methyltransferase
J.Med.Chem., 49, 2006
|
|
3F78
 
 | | Crystal structure of wild type LFA1 I domain complexed with isoflurane | | 分子名称: | 1-CHLORO-2,2,2-TRIFLUOROETHYL DIFLUOROMETHYL ETHER, GLYCEROL, Integrin alpha-L, ... | | 著者 | Zhang, H, Wang, J.-H. | | 登録日 | 2008-11-07 | | 公開日 | 2009-06-23 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structure of isoflurane bound to integrin LFA-1 supports a unified mechanism of volatile anesthetic action in the immune and central nervous systems.
Faseb J., 23, 2009
|
|
1EX5
 
 | |
1EYN
 
 | | Structure of mura liganded with the extrinsic fluorescence probe ANS | | 分子名称: | 8-ANILINO-1-NAPHTHALENE SULFONATE, GLYCEROL, UDP-N-ACETYLGLUCOSAMINE 1-CARBOXYVINYLTRANSFERASE | | 著者 | Schonbrunn, E, Eschenburg, S, Luger, K, Kabsch, W, Amrhein, N. | | 登録日 | 2000-05-07 | | 公開日 | 2000-06-09 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural basis for the interaction of the fluorescence probe 8-anilino-1-naphthalene sulfonate (ANS) with the antibiotic target MurA.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1RED
 
 | | ENDO-1,4-BETA-XYLANASE II COMPLEX WITH 4,5-EPOXYPENTYL-BETA-D-XYLOSIDE | | 分子名称: | 3-[(2R)-oxiran-2-yl]propyl beta-D-xylopyranoside, BENZOIC ACID, ENDO-1,4-BETA-XYLANASE II | | 著者 | Rouvinen, J, Havukainen, R, Torronen, A. | | 登録日 | 1995-12-21 | | 公開日 | 1997-01-11 | | 最終更新日 | 2024-12-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Covalent binding of three epoxyalkyl xylosides to the active site of endo-1,4-xylanase II from Trichoderma reesei.
Biochemistry, 35, 1996
|
|
3D61
 
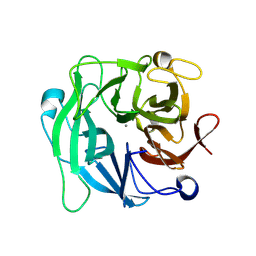 | | Crystal Structure Analysis of 1,5-alpha-arabinanase catalytic mutant (AbnBD147A) complexed to arabinobiose | | 分子名称: | CALCIUM ION, Intracellular arabinanase, alpha-L-arabinofuranose-(1-5)-beta-L-arabinofuranose | | 著者 | Alhassid, A, Ben David, A, Shoham, Y, Shoham, G. | | 登録日 | 2008-05-18 | | 公開日 | 2009-04-21 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure of an inverting GH 43 1,5-alpha-L-arabinanase from Geobacillus stearothermophilus complexed with its substrate
Biochem.J., 422, 2009
|
|
4OQB
 
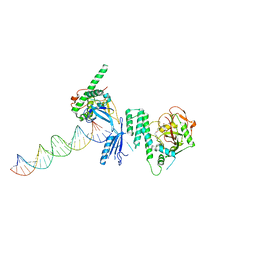 | | Structure of Human PARP-1 bound to a DNA double strand break in complex with (2Z)-2-{4-[2-(morpholin-4-yl)ethoxy]benzylidene}-3-oxo-2,3-dihydro-1-benzofuran-7-carboxamide | | 分子名称: | (2Z)-2-{4-[2-(morpholin-4-yl)ethoxy]benzylidene}-3-oxo-2,3-dihydro-1-benzofuran-7-carboxamide, DNA (26-MER), Poly [ADP-ribose] polymerase 1, ... | | 著者 | Pascal, J.M, Steffen, J.D. | | 登録日 | 2014-02-07 | | 公開日 | 2014-07-02 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.362 Å) | | 主引用文献 | Discovery and Structure-Activity Relationship of Novel 2,3-Dihydrobenzofuran-7-carboxamide and 2,3-Dihydrobenzofuran-3(2H)-one-7-carboxamide Derivatives as Poly(ADP-ribose)polymerase-1 Inhibitors.
J.Med.Chem., 57, 2014
|
|
3D5Z
 
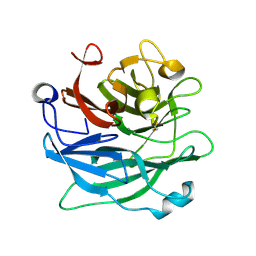 | | Crystal Structure Analysis of 1,5-alpha-arabinanase catalytic mutant (AbnBE201A) complexed to arabinotriose | | 分子名称: | CALCIUM ION, Intracellular arabinanase, alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-beta-L-arabinofuranose | | 著者 | Alhassid, A, Ben David, A, Shoham, Y, Shoham, G. | | 登録日 | 2008-05-18 | | 公開日 | 2009-04-21 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of an inverting GH 43 1,5-alpha-L-arabinanase from Geobacillus stearothermophilus complexed with its substrate
Biochem.J., 422, 2009
|
|
