7YK6
 
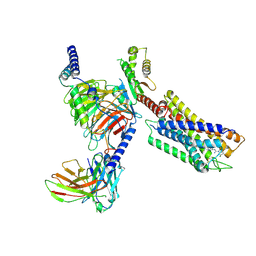 | | Cryo-EM structure of the compound 4-bound human relaxin family peptide receptor 4 (RXFP4)-Gi complex | | 分子名称: | 1-[2-(4-chlorophenyl)ethyl]-3-[(7-ethyl-5-oxidanyl-1H-indol-3-yl)methylideneamino]guanidine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Chen, Y, Zhou, Q.T, Wang, J, Xu, Y.W, Wang, Y, Yan, J.H, Wang, Y.B, Zhu, Q, Zhao, F.H, Li, C.H, Chen, C.W, Cai, X.Q, Bathgate, R.A.D, Shen, C, Liu, H, Xu, H.E, Yang, D.H, Wang, M.W. | | 登録日 | 2022-07-21 | | 公開日 | 2023-03-01 | | 最終更新日 | 2025-06-25 | | 実験手法 | ELECTRON MICROSCOPY (3.03 Å) | | 主引用文献 | Ligand recognition mechanism of the human relaxin family peptide receptor 4 (RXFP4).
Nat Commun, 14, 2023
|
|
2XTK
 
 | |
7GD5
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ALP-POS-d2866bdf-1 (Mpro-x10876) | | 分子名称: | 2-(1H-benzotriazol-1-yl)-N-[4-(dimethylamino)phenyl]-N-[(thiophen-3-yl)methyl]acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.809 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
6FE7
 
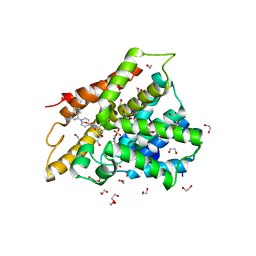 | | Crystal structure of human phosphodiesterase 4D2 catalytic domain with inhibitor NPD-356 | | 分子名称: | (4aS,8aR)-2-(1-{2-aminothieno[2,3-d]pyrimidin-4-yl}piperidin-4-yl)-4-(3,4- dimethoxyphenyl)-1,2,4a,5,8,8a-hexahydrophthalazin-1-one, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | 著者 | Singh, A.K, Brown, D.G. | | 登録日 | 2017-12-29 | | 公開日 | 2019-04-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | hPDE4D2 structure with inhibitor NPD-356
To be published
|
|
4G75
 
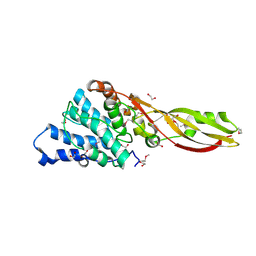 | | Structure of PaeM, a colicin M-like bacteriocin produced by Pseudomonas aeruginosa | | 分子名称: | 1,2-ETHANEDIOL, MAGNESIUM ION, phosphodiesterase | | 著者 | Touze, T, Graille, M, Mengin-Lecreulx, D. | | 登録日 | 2012-07-20 | | 公開日 | 2012-09-12 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Functional and Structural Characterization of PaeM, a Colicin M-like Bacteriocin Produced by Pseudomonas aeruginosa.
J.Biol.Chem., 287, 2012
|
|
6QTZ
 
 | |
6B4D
 
 | | Crystal structure of human carbonic anhydrase II in complex with a heteroaryl-pyrazole carboxylic acid derivative. | | 分子名称: | 3-(1-ethyl-1H-indol-3-yl)-1-methyl-1H-pyrazole-5-carboxylic acid, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | 著者 | Lomelino, C.L, Mahon, B.P, McKenna, R. | | 登録日 | 2017-09-26 | | 公開日 | 2018-02-07 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.196 Å) | | 主引用文献 | Exploring Heteroaryl-pyrazole Carboxylic Acids as Human Carbonic Anhydrase XII Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
9MIN
 
 | |
7GE2
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDJ-MED-6af13d92-1 (Mpro-x11313) | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[2-(2-methoxyphenoxy)ethyl]-5-methyl-2-oxo-1,2-dihydroquinoline-4-carboxamide | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
5DZ8
 
 | | Streptococcus agalactiae AgI/II polypeptide BspA variable (V) domain | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, BspA (BspA_V), ... | | 著者 | Rego, S, Till, M, Race, P.R. | | 登録日 | 2015-09-25 | | 公開日 | 2016-06-22 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Structural and Functional Analysis of Cell Wall-anchored Polypeptide Adhesin BspA in Streptococcus agalactiae.
J.Biol.Chem., 291, 2016
|
|
7GNG
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDJ-MED-705e09b8-1 (Mpro-P2607) | | 分子名称: | 2-[(3'S)-6-chloro-2'-oxo-1'-(5,6,7,8-tetrahydroisoquinolin-4-yl)-1H-spiro[isoquinoline-4,3'-pyrrolidin]-2(3H)-yl]-N-methylacetamide, 3C-like proteinase, CHLORIDE ION, ... | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.769 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
8ITU
 
 | | SARS-CoV-2 Omicron BA.1 Spike glycoprotein in complex with rabbit monoclonal antibody 1H1 IgG. | | 分子名称: | 1H1 heavy chain, 1H1 light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | 登録日 | 2023-03-23 | | 公開日 | 2023-04-12 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (3.68 Å) | | 主引用文献 | Mechanism of a rabbit monoclonal antibody broadly neutralizing SARS-CoV-2 variants.
Commun Biol, 6, 2023
|
|
7GI3
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-090737b9-1 (Mpro-P0041) | | 分子名称: | (4R)-6-chloro-N-(isoquinolin-4-yl)-N-propanoyl-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, CALCIUM ION, ... | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
6SY7
 
 | | Structure of Trypanosome Brucei Phosphofructokinase in complex with AMP. | | 分子名称: | ADENOSINE MONOPHOSPHATE, ATP-dependent 6-phosphofructokinase, BENZENE, ... | | 著者 | McNae, I.W, Vasquez-Valdivieso, M.G, Walkinshaw, M.D. | | 登録日 | 2019-09-27 | | 公開日 | 2019-12-25 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Kinetic and structural studies of Trypanosoma and Leishmania phosphofructokinases show evolutionary divergence and identify AMP as a switch regulating glycolysis versus gluconeogenesis.
Febs J., 287, 2020
|
|
9D4E
 
 | | Crystal structure of the human DCAF1 WDR domain in complex with OICR-41103 | | 分子名称: | (4P)-N-[(1S)-3-amino-1-(3-chloro-4-fluorophenyl)-3-oxopropyl]-4-(4-chloro-2-fluorophenyl)-5-[(1E)-3-oxo-3-(piperidin-1-yl)prop-1-en-1-yl]-1H-pyrrole-3-carboxamide, CITRIC ACID, DDB1- and CUL4-associated factor 1 | | 著者 | Kimani, S, Dong, A, Li, Y, Seitova, A, Al-awar, R, Mamai, A, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | 登録日 | 2024-08-12 | | 公開日 | 2024-12-18 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of the human DCAF1 WDR domain in complex with OICR-1103
To be published
|
|
6SC4
 
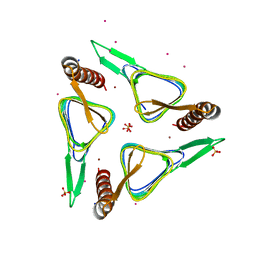 | | Gamma-Carbonic Anhydrase from the Haloarchaeon Halobacterium sp. | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CADMIUM ION, ... | | 著者 | Vogler, M, Karan, R, Renn, D, Vancea, A, Vielberg, V.-T, Groetzinger, S.W, DasSarma, P, Das Sarma, S, Eppinger, J, Groll, M, Rueping, M. | | 登録日 | 2019-07-23 | | 公開日 | 2020-04-22 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal Structure and Active Site Engineering of a Halophilic gamma-Carbonic Anhydrase.
Front Microbiol, 11, 2020
|
|
6MAE
 
 | |
8A1H
 
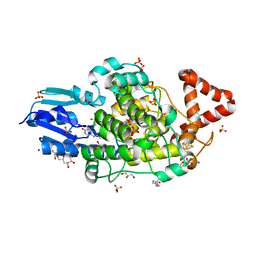 | | Bacterial 6-4 photolyase from Vibrio cholerase | | 分子名称: | 1-deoxy-1-(6,7-dimethyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 6-4 photolyase (FeS-BCP, ... | | 著者 | Essen, L.-O, Emmerich, H.J. | | 登録日 | 2022-06-01 | | 公開日 | 2023-02-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural and Functional Analysis of a Prokaryotic (6-4) Photolyase from the Aquatic Pathogen Vibrio Cholerae † .
Photochem.Photobiol., 99, 2023
|
|
7GKC
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ALP-POS-9c80c481-1 (Mpro-P0845) | | 分子名称: | (4R)-6-chloro-N-(isoquinolin-4-yl)-4-{[2-(methylamino)-2-oxoethoxy]methyl}-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GL5
 
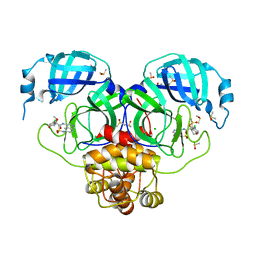 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDG-MED-10fcb19e-1 (Mpro-P1661) | | 分子名称: | (4S)-6-chloro-N-{6-[(methanesulfonyl)amino]isoquinolin-4-yl}-4-methoxy-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.678 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
4GFT
 
 | | Malaria invasion machinery protein-Nanobody complex | | 分子名称: | 1,2-ETHANEDIOL, Myosin A tail domain interacting protein, Nanobody | | 著者 | Khamrui, S, Turley, S, Pardon, E, Steyaert, J, Verlinde, C, Fan, E, Bergman, L.W, Hol, W.G.J. | | 登録日 | 2012-08-03 | | 公開日 | 2013-07-03 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The structure of the D3 domain of Plasmodium falciparum myosin tail interacting protein MTIP in complex with a nanobody.
Mol.Biochem.Parasitol., 190, 2013
|
|
1LA2
 
 | | Structural analysis of Saccharomyces cerevisiae myo-inositol phosphate synthase | | 分子名称: | Myo-inositol-1-phosphate synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Kniewel, R, Buglino, J.A, Shen, V, Chadna, T, Beckwith, A, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2002-03-27 | | 公開日 | 2002-04-10 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structural analysis of Saccharomyces cerevisiae myo-inositol phosphate synthase
J.STRUCT.FUNCT.GENOM., 2, 2002
|
|
1D2I
 
 | | CRYSTAL STRUCTURE OF RESTRICTION ENDONUCLEASE BGLII COMPLEXED WITH DNA 16-MER | | 分子名称: | DNA (5'-D(*TP*AP*TP*TP*AP*TP*AP*GP*AP*TP*CP*TP*AP*TP*AP*A)-3'), MAGNESIUM ION, PROTEIN (RESTRICTION ENDONUCLEASE BGLII) | | 著者 | Lukacs, C.M, Kucera, R, Schildkraut, I, Aggarwal, A.K. | | 登録日 | 1999-09-23 | | 公開日 | 2000-02-21 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Understanding the immutability of restriction enzymes: crystal structure of BglII and its DNA substrate at 1.5 A resolution.
Nat.Struct.Biol., 7, 2000
|
|
7UDL
 
 | | Crystal structure of designed helical repeat protein RPB_PLP1_R6 bound to PLPx6 peptide | | 分子名称: | 1,2-ETHANEDIOL, 6xPLP Peptide, Designed helical repeat protein (DHR) RPB_PLP1_R6 | | 著者 | Chang, Y, Redler, R.L, Bhabha, G, Ekiert, D.C. | | 登録日 | 2022-03-20 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
6BAI
 
 | | Multiconformer model of apo K197C PTP1B at 100 K | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | 著者 | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | 登録日 | 2017-10-12 | | 公開日 | 2018-06-20 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
