6FIQ
 
 | | DDR1, 1-(1H-indazole-5-carbonyl)-5'-methoxy-1'-[2-oxo-2-[(2S)-2-(trifluoromethyl)pyrrolidin-1-yl]ethyl]spiro[piperidine-4,3'-pyrrolo[3,2-b]pyridine]-2'-one, 1.790A, P212121, Rfree=23.8% | | 分子名称: | 1-(1~{H}-indazol-5-ylcarbonyl)-5'-methoxy-1'-[2-oxidanylidene-2-[(2~{S})-2-(trifluoromethyl)pyrrolidin-1-yl]ethyl]spiro[piperidine-4,3'-pyrrolo[3,2-b]pyridine]-2'-one, CHLORIDE ION, Epithelial discoidin domain-containing receptor 1 | | 著者 | Stihle, M, Richter, H, Benz, J, Kuhn, B, Rudolph, M.G. | | 登録日 | 2018-01-19 | | 公開日 | 2018-11-28 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | DNA-Encoded Library-Derived DDR1 Inhibitor Prevents Fibrosis and Renal Function Loss in a Genetic Mouse Model of Alport Syndrome.
Acs Chem.Biol., 14, 2019
|
|
6BTO
 
 | | BMP1 complexed with (2~{S})-2-[[(1~{R},3~{S},4~{S})-2-[(2~{R})-2-[2-(oxidanylamino)-2-oxidanylidene-ethyl]heptanoyl]-2-azabicyclo[2.2.1]heptan-3-yl]carbonylamino]-2-phenyl-ethanoic acid | | 分子名称: | (2~{S})-2-[[(1~{R},3~{S},4~{S})-2-[(2~{R})-2-[2-(oxidanylamino)-2-oxidanylidene-ethyl]heptanoyl]-2-azabicyclo[2.2.1]heptan-3-yl]carbonylamino]-2-phenyl-ethanoic acid, 1,2-ETHANEDIOL, Bone morphogenetic protein 1, ... | | 著者 | Gampe, R, Shewchuk, L. | | 登録日 | 2017-12-07 | | 公開日 | 2018-08-08 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Reverse Hydroxamate Inhibitors of Bone Morphogenetic Protein 1.
ACS Med Chem Lett, 9, 2018
|
|
7KEP
 
 | | avibactam-CDD-1 2 minute complex | | 分子名称: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, (2S,5R)-7-oxo-6-(sulfooxy)-1,6-diazabicyclo[3.2.1]octane-2-carboxamide, (4S)-2-METHYL-2,4-PENTANEDIOL, ... | | 著者 | Smith, C.A, Vakulenko, S.B, Stewart, N.K, Toth, M. | | 登録日 | 2020-10-11 | | 公開日 | 2021-01-20 | | 最終更新日 | 2025-04-02 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Inhibition of the Clostridioides difficile Class D beta-Lactamase CDD-1 by Avibactam.
Acs Infect Dis., 7, 2021
|
|
7Y8W
 
 | | Crystal structure of DLC-1/SAO-1 complex | | 分子名称: | Dynein light chain 1, cytoplasmic, GLYCEROL, ... | | 著者 | Yan, H, Zhao, C, Wei, Z, Yu, C. | | 登録日 | 2022-06-24 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Interaction between DLC-1 and SAO-1 facilitates CED-4 translocation during apoptosis in the Caenorhabditis elegans germline.
Cell Death Discov, 8, 2022
|
|
2RF5
 
 | | Crystal structure of human tankyrase 1- catalytic PARP domain | | 分子名称: | GLYCEROL, Tankyrase-1, ZINC ION | | 著者 | Lehtio, L, Karlberg, T, Arrowsmith, C.H, Berglund, H, Busam, R, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Herman, M.D, Holmberg-Schiavone, L, Johansson, I, Kallas, A, Kotenyova, T, Moche, M, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Sundstrom, M, Thorsell, A.G, Tresaugues, L, van den Berg, S, Welin, M, Weigelt, J, Structural Genomics Consortium (SGC) | | 登録日 | 2007-09-28 | | 公開日 | 2007-10-09 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Zinc binding catalytic domain of human tankyrase 1.
J.Mol.Biol., 379, 2008
|
|
5I92
 
 | |
1LV1
 
 | | Crystal Structure Analysis of the non-active site mutant of tethered HIV-1 protease to 2.1A resolution | | 分子名称: | HIV-1 protease | | 著者 | Kumar, M, Kannan, K.K, Hosur, M.V, Bhavesh, N.S, Chatterjee, A, Mittal, R, Hosur, R.V. | | 登録日 | 2002-05-24 | | 公開日 | 2002-06-19 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Effects of remote mutation on the autolysis of HIV-1 PR: X-ray and NMR investigations.
Biochem.Biophys.Res.Commun., 294, 2002
|
|
4BZT
 
 | | The Solution Structure of the MLN 944-d(ATGCAT)2 Complex | | 分子名称: | 1-METHYL-9-[12-(9-METHYLPHENAZIN-10-IUM-1-YL)-12-OXO-2,11-DIAZA-5,8-DIAZONIADODEC-1-ANOYL]PHENAZIN-10-IUM, DNA | | 著者 | Serobian, A, Thomas, D.S, Ball, G.E, Denny, W.A, Wakelin, L.P.G. | | 登録日 | 2013-07-30 | | 公開日 | 2013-08-21 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Solution Structure of Bis(Phenazine-1-Carboxamide)-DNA Complexes: Mln 944 Binding Corrected and Extended.
Biopolymers, 101, 2014
|
|
5ZGE
 
 | | Crystal structure of NDM-1 at pH5.5 (Bis-Tris) in complex with hydrolyzed ampicillin | | 分子名称: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, HYDROXIDE ION, Metallo-beta-lactamase type 2, ... | | 著者 | Zhang, H, Hao, Q. | | 登録日 | 2018-03-08 | | 公開日 | 2018-08-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Active-Site Conformational Fluctuations Promote the Enzymatic Activity of NDM-1.
Antimicrob. Agents Chemother., 62, 2018
|
|
5EPC
 
 | |
7YMG
 
 | | Crystal structure of BRD4 bromodomain 1 (BD1) in complex with 2-({3-ethyl-[1,2,4]triazolo[4,3-b]pyridazin-6-yl}amino)-3-(1H-indol-3-yl)propan-1-ol | | 分子名称: | (2S)-2-[(3-ethyl-[1,2,4]triazolo[4,3-b]pyridazin-6-yl)amino]-3-(1H-indol-3-yl)propan-1-ol, Bromodomain-containing protein 4, FORMIC ACID, ... | | 著者 | Kim, J.H, Lee, B.I. | | 登録日 | 2022-07-28 | | 公開日 | 2023-01-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal structure of [1,2,4]triazolo[4,3-b]pyridazine derivatives as BRD4 bromodomain inhibitors and structure-activity relationship study.
Sci Rep, 13, 2023
|
|
5J8O
 
 | | Structure of human Programmed cell death 1 ligand 1 (PD-L1) with low molecular mass inhibitor | | 分子名称: | (2R)-1-({3-bromo-4-[(2-methyl[1,1'-biphenyl]-3-yl)methoxy]phenyl}methyl)piperidine-2-carboxylic acid, Programmed cell death 1 ligand 1 | | 著者 | Zak, K.M, Grudnik, P, Guzik, K, Zieba, B.J, Musielak, B, Doemling, P, Dubin, G, Holak, T.A. | | 登録日 | 2016-04-08 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis for small molecule targeting of the programmed death ligand 1 (PD-L1).
Oncotarget, 7, 2016
|
|
6LLF
 
 | | Biphenyl-2,2',3-triol-soaked resting complex of Oxy and Fd in carbazole 1,9a-dioxygenase | | 分子名称: | 1,2-ETHANEDIOL, 3-(2-hydroxyphenyl)benzene-1,2-diol, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Wang, Y.X, Suzuki-Minakuchi, C, Nojiri, H. | | 登録日 | 2019-12-23 | | 公開日 | 2021-01-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Biphenyl-2,2',3-triol-soaked resting complex of Oxy and Fd in carbazole 1,9a-dioxygenase
To Be Published
|
|
9GCQ
 
 | | Human Butyrylcholinesterase in complex with N1,N1-dimethyl-N2-(6-(naphthalen-2-yl)-5-(pyridin-4-yl)pyridazin-3-yl)ethane-1,2-diamine | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Brazzolotto, X, Meden, A, Knez, D, Gobec, S, Nachon, F. | | 登録日 | 2024-08-02 | | 公開日 | 2025-08-13 | | 実験手法 | X-RAY DIFFRACTION (2.62 Å) | | 主引用文献 | Human Butyrylcholinesterase in complex with N1,N1-dimethyl-N2-(6-(naphthalen-2-yl)-5-(pyridin-4-yl)pyridazin-3-yl)ethane-1,2-diamine
To Be Published
|
|
6FS1
 
 | | MCL1 in complex with an indole acid ligand | | 分子名称: | 1,2-ETHANEDIOL, 7-[3-[(1,5-dimethylpyrazol-3-yl)methylsulfanylmethyl]-1,5-dimethyl-pyrazol-4-yl]-3-(3-naphthalen-1-yloxypropyl)-1~{H}-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | 著者 | Kasmirski, S, Hargreaves, D. | | 登録日 | 2018-02-18 | | 公開日 | 2018-12-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Discovery of Mcl-1-specific inhibitor AZD5991 and preclinical activity in multiple myeloma and acute myeloid leukemia.
Nat Commun, 9, 2018
|
|
1ZKN
 
 | | Structure of PDE4D2-IBMX | | 分子名称: | 3-ISOBUTYL-1-METHYLXANTHINE, MAGNESIUM ION, ZINC ION, ... | | 著者 | Huai, Q, Liu, Y, Francis, S.H, Corbin, J.D, Ke, H. | | 登録日 | 2005-05-03 | | 公開日 | 2005-05-17 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structures of Phosphodiesterases 4 and 5 in Complex with Inhibitor 3-Isobutyl-1-Methylxanthine Suggest a Conformation Determinant of Inhibitor Selectivity
J.Biol.Chem., 279, 2004
|
|
4YHQ
 
 | | Crystal structure of multidrug resistant clinical isolate PR20 with GRL-5010A | | 分子名称: | (3R,3aS,6aS)-4,4-difluorohexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{[(4-aminophenyl)sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, GLYCEROL, ... | | 著者 | Agniswamy, J, Weber, I.T. | | 登録日 | 2015-02-27 | | 公開日 | 2015-06-10 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Substituted Bis-THF Protease Inhibitors with Improved Potency against Highly Resistant Mature HIV-1 Protease PR20.
J.Med.Chem., 58, 2015
|
|
1YDR
 
 | | STRUCTURE OF CAMP-DEPENDENT PROTEIN KINASE, ALPHA-CATALYTIC SUBUNIT IN COMPLEX WITH H7 PROTEIN KINASE INHIBITOR 1-(5-ISOQUINOLINESULFONYL)-2-METHYLPIPERAZINE | | 分子名称: | 1-(5-ISOQUINOLINESULFONYL)-2-METHYLPIPERAZINE, C-AMP-DEPENDENT PROTEIN KINASE, PROTEIN KINASE INHIBITOR PEPTIDE | | 著者 | Engh, R.A, Girod, A, Kinzel, V, Huber, R, Bossemeyer, D. | | 登録日 | 1996-07-24 | | 公開日 | 1997-04-01 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structures of catalytic subunit of cAMP-dependent protein kinase in complex with isoquinolinesulfonyl protein kinase inhibitors H7, H8, and H89. Structural implications for selectivity.
J.Biol.Chem., 271, 1996
|
|
2I0Y
 
 | |
6AYA
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with Nup153 peptide | | 分子名称: | CHLORIDE ION, HIV-1 capsid protein, IODIDE ION, ... | | 著者 | Gres, A.T, Kirby, K.A, Sarafianos, S.G. | | 登録日 | 2017-09-07 | | 公開日 | 2018-09-12 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Multidisciplinary studies with mutated HIV-1 capsid proteins reveal structural mechanisms of lattice stabilization.
Nat Commun, 14, 2023
|
|
5CDP
 
 | | 2.45A structure of etoposide with S.aureus DNA gyrase and DNA | | 分子名称: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*G*GP*GP*TP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), DNA gyrase subunit A, ... | | 著者 | Bax, B.D, Srikannathasan, V, Chan, P.F. | | 登録日 | 2015-07-04 | | 公開日 | 2015-12-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
4ZYF
 
 | |
7JN0
 
 | | Sheep Connexin-46 at 2.5 angstroms resolution, Lipid Class 2 | | 分子名称: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-3 protein | | 著者 | Flores, J.A, Haddad, B.G, Dolan, K.A, Myers, J.A, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | 登録日 | 2020-08-03 | | 公開日 | 2020-09-09 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
7YCC
 
 | | KRas G12C in complex with Compound 5c | | 分子名称: | 1-[7-[6-chloranyl-8-fluoranyl-7-(5-methyl-1~{H}-indazol-4-yl)-2-[(1-methylpiperidin-4-yl)amino]quinazolin-4-yl]-2,7-diazaspiro[3.5]nonan-2-yl]propan-1-one, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | 著者 | Amano, Y. | | 登録日 | 2022-07-01 | | 公開日 | 2022-08-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Discovery and biological evaluation of 1-{2,7-diazaspiro[3.5]nonan-2-yl}prop-2-en-1-one derivatives as covalent inhibitors of KRAS G12C with favorable metabolic stability and anti-tumor activity.
Bioorg.Med.Chem., 71, 2022
|
|
6BX7
 
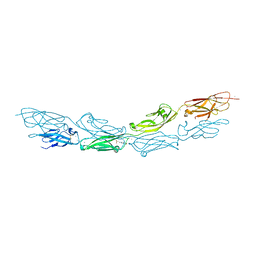 | | Crystal Structure of Human Protocadherin-1 EC1-4 | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, Protocadherin-1 | | 著者 | Modak, D, Sotomayor, M. | | 登録日 | 2017-12-17 | | 公開日 | 2018-12-19 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Identification of an adhesive interface for the non-clustered delta 1 protocadherin-1 involved in respiratory diseases.
Commun Biol, 2, 2019
|
|
