7DOI
 
 | |
7YSX
 
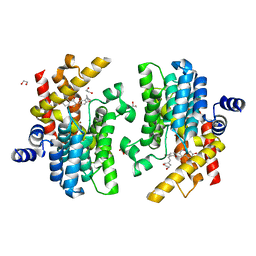 | | Crystal structure of PDE4D complexed with licoisoflavone A | | 分子名称: | 1,2-ETHANEDIOL, 3-[3-(3-methylbut-2-enyl)-2,4-bis(oxidanyl)phenyl]-5,7-bis(oxidanyl)chromen-4-one, MAGNESIUM ION, ... | | 著者 | Liu, J.Y, Li, M.J, Xu, Y.C. | | 登録日 | 2022-08-13 | | 公開日 | 2023-07-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Bioactive compounds from Huashi Baidu decoction possess both antiviral and anti-inflammatory effects against COVID-19.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7YQF
 
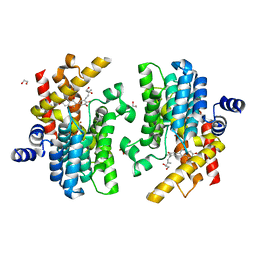 | | Crystal structure of PDE4D complexed with glycyrrhisoflavone | | 分子名称: | 1,2-ETHANEDIOL, 3-[3-(3-methylbut-2-enyl)-4,5-bis(oxidanyl)phenyl]-5,7-bis(oxidanyl)chromen-4-one, MAGNESIUM ION, ... | | 著者 | Liu, J.Y, Li, M.J, Xu, Y.C. | | 登録日 | 2022-08-06 | | 公開日 | 2023-07-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Bioactive compounds from Huashi Baidu decoction possess both antiviral and anti-inflammatory effects against COVID-19.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6YUN
 
 | |
6W63
 
 | |
6ZCO
 
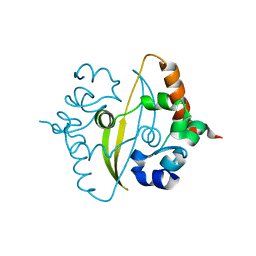 | | Crystal Structure of C-terminal Dimerization Domain of Nucleocapsid Phosphoprotein from SARS-CoV-2, crystal form II | | 分子名称: | Nucleoprotein | | 著者 | Zinzula, L, Basquin, J, Nagy, I, Bracher, A. | | 登録日 | 2020-06-11 | | 公開日 | 2020-07-01 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.361 Å) | | 主引用文献 | High-resolution structure and biophysical characterization of the nucleocapsid phosphoprotein dimerization domain from the Covid-19 severe acute respiratory syndrome coronavirus 2.
Biochem.Biophys.Res.Commun., 538, 2021
|
|
6ZRT
 
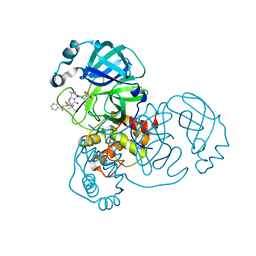 | | Crystal structure of SARS CoV2 main protease in complex with inhibitor Telaprevir | | 分子名称: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, DIMETHYL SULFOXIDE, Main Protease | | 著者 | Oerlemans, R, Wang, W, Lunev, S, Domling, A, Groves, M.R. | | 登録日 | 2020-07-14 | | 公開日 | 2020-08-12 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Repurposing the HCV NS3-4A protease drug boceprevir as COVID-19 therapeutics.
Rsc Med Chem, 12, 2020
|
|
6ZRU
 
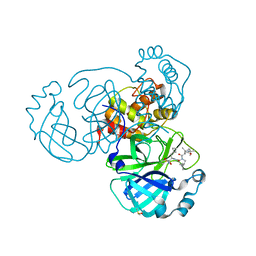 | | Crystal structure of SARS CoV2 main protease in complex with inhibitor Boceprevir | | 分子名称: | DIMETHYL SULFOXIDE, Main Protease, boceprevir (bound form) | | 著者 | Oerlemans, R, Wang, W, Lunev, S, Domling, A, Groves, M.R. | | 登録日 | 2020-07-14 | | 公開日 | 2020-08-12 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Repurposing the HCV NS3-4A protease drug boceprevir as COVID-19 therapeutics.
Rsc Med Chem, 12, 2020
|
|
7EIN
 
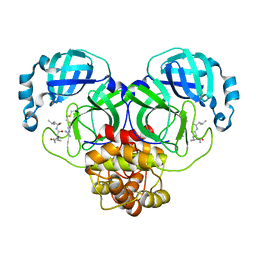 | | SARS-CoV-2 main proteinase complex with microbial metabolite leupeptin | | 分子名称: | 3C-like proteinase, leupeptin | | 著者 | Fu, L.F, Feng, Y, Qi, J.X, Gao, G.F. | | 登録日 | 2021-03-31 | | 公開日 | 2021-07-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Mechanism of Microbial Metabolite Leupeptin in the Treatment of COVID-19 by Traditional Chinese Medicine Herbs.
Mbio, 12, 2021
|
|
7OAQ
 
 | |
7OAO
 
 | | Nanobody C5 bound to RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, C5 nanobody, ... | | 著者 | Naismith, J.H, Mikolajek, H. | | 登録日 | 2021-04-19 | | 公開日 | 2021-08-11 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19.
Nat Commun, 12, 2021
|
|
7OAP
 
 | | Nanobody H3 AND C1 bound to RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C1 nanobody, CHLORIDE ION, ... | | 著者 | Naismith, J.H, Mikolajek, H. | | 登録日 | 2021-04-19 | | 公開日 | 2021-08-11 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.901 Å) | | 主引用文献 | A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19.
Nat Commun, 12, 2021
|
|
7OAN
 
 | | Nanobody C5 bound to Spike | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Naismith, J.H, Weckener, M. | | 登録日 | 2021-04-19 | | 公開日 | 2021-08-11 | | 最終更新日 | 2021-10-06 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19.
Nat Commun, 12, 2021
|
|
7OAY
 
 | | Nanobody F2 bound to RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F2 nanobody, Spike protein S1 | | 著者 | Naismith, J.H, Mikolajek, H. | | 登録日 | 2021-04-20 | | 公開日 | 2021-08-11 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19.
Nat Commun, 12, 2021
|
|
7OAU
 
 | |
8FTL
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease (Mpro) in complex with inhibitor Jun89-3-C1 | | 分子名称: | 3C-like proteinase nsp5, N-([1,1'-biphenyl]-4-yl)-2-chloro-N-[(1R)-2-oxo-2-{[(1S)-1-phenylethyl]amino}-1-(pyridin-3-yl)ethyl]acetamide | | 著者 | Lewandowski, E.M, Butler, S.G, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2023-01-12 | | 公開日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Crystal structure of the SARS-CoV-2 (COVID-19) main protease (Mpro) in complex with inhibitor Jun89-3-C1
To Be Published
|
|
6W4B
 
 | | The crystal structure of Nsp9 RNA binding protein of SARS CoV-2 | | 分子名称: | Non-structural protein 9 | | 著者 | Tan, K, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-03-10 | | 公開日 | 2020-03-18 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | The crystal structure of Nsp9 replicase protein of COVID-19
To Be Published
|
|
7E7B
 
 | | Cryo-EM structure of the SARS-CoV-2 furin site mutant S-Trimer from a subunit vaccine candidate | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-hydroxyethyl 2-deoxy-3,5-bis-O-(2-hydroxyethyl)-6-O-(2-{[(9E)-octadec-9-enoyl]oxy}ethyl)-alpha-L-xylo-hexofuranoside, ... | | 著者 | Zheng, S, Ma, J. | | 登録日 | 2021-02-25 | | 公開日 | 2021-03-24 | | 最終更新日 | 2022-12-07 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Cryo-EM structure of S-Trimer, a subunit vaccine candidate for COVID-19.
J.Virol., 95, 2021
|
|
7E7D
 
 | |
8X0X
 
 | | Crystal structure of JE-5C in complex with SARS-CoV-2 RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of JE-5C Fab, Light chain of JE-5C Fab, ... | | 著者 | Mohapatra, A, Chen, X. | | 登録日 | 2023-11-06 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (3.45 Å) | | 主引用文献 | The presence of broadly neutralizing anti-SARS-CoV-2 RBD antibodies elicited by primary series and booster dose of COVID-19 vaccine.
Plos Pathog., 20, 2024
|
|
8YRP
 
 | | SARS-CoV-2 Delta Spike in complex with JM-1A | | 分子名称: | JM-1A Heavy Chain, JM-1A Light Chain, Spike glycoprotein | | 著者 | Nguyen, V.H.T, Chen, X. | | 登録日 | 2024-03-21 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.64 Å) | | 主引用文献 | The presence of broadly neutralizing anti-SARS-CoV-2 RBD antibodies elicited by primary series and booster dose of COVID-19 vaccine.
Plos Pathog., 20, 2024
|
|
8X0Y
 
 | | Crystal structure of JM-1A in complex with SARS-CoV-2 RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Heavy chain of JM-1A Fab, ... | | 著者 | Mohapatra, A, Chen, X. | | 登録日 | 2023-11-06 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | The presence of broadly neutralizing anti-SARS-CoV-2 RBD antibodies elicited by primary series and booster dose of COVID-19 vaccine.
Plos Pathog., 20, 2024
|
|
8YRO
 
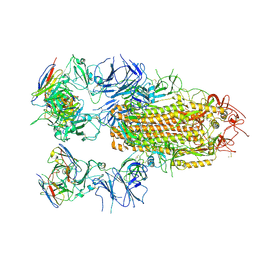 | | SARS-CoV-2 Delta Spike in complex with JL-8C | | 分子名称: | JL-8C Heavy Chain, JL-8C Light Chain, Spike glycoprotein | | 著者 | Nguyen, V.H.T, Chen, X. | | 登録日 | 2024-03-21 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.27 Å) | | 主引用文献 | The presence of broadly neutralizing anti-SARS-CoV-2 RBD antibodies elicited by primary series and booster dose of COVID-19 vaccine.
Plos Pathog., 20, 2024
|
|
8YZ5
 
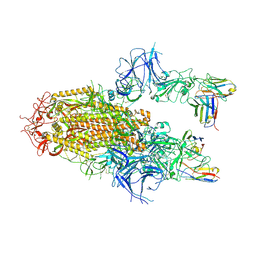 | | SARS-CoV-2 Delta Spike in complex with Fab of JE-5C | | 分子名称: | Fab heavy chain of JE-5C, Fab light chain of JE-5C, Spike glycoprotein | | 著者 | Chen, X, Wu, Y.-M. | | 登録日 | 2024-04-06 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.93 Å) | | 主引用文献 | The presence of broadly neutralizing anti-SARS-CoV-2 RBD antibodies elicited by primary series and booster dose of COVID-19 vaccine.
Plos Pathog., 20, 2024
|
|
8YZ6
 
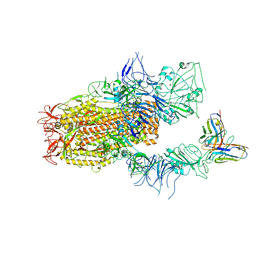 | | SARS-CoV-2 Spike (BA.1) in complex with Fab of JH-8B | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab heavy chain of JH-8B, ... | | 著者 | Chen, X, Wu, Y.-M. | | 登録日 | 2024-04-06 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (4.55 Å) | | 主引用文献 | The presence of broadly neutralizing anti-SARS-CoV-2 RBD antibodies elicited by primary series and booster dose of COVID-19 vaccine.
Plos Pathog., 20, 2024
|
|
