7S36
 
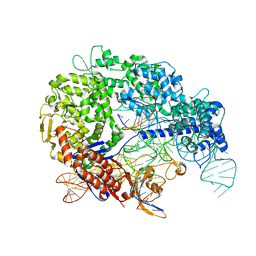 | | Cas9:sgRNA:DNA (S. pyogenes) with 0 RNA:DNA base pairs, closed-protein/bent-DNA conformation | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1, Non-target DNA strand, Single-guide RNA, ... | | 著者 | Cofsky, J.C, Soczek, K.M, Knott, G.J, Nogales, E, Doudna, J.A. | | 登録日 | 2021-09-04 | | 公開日 | 2022-04-20 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | CRISPR-Cas9 bends and twists DNA to read its sequence.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7S3H
 
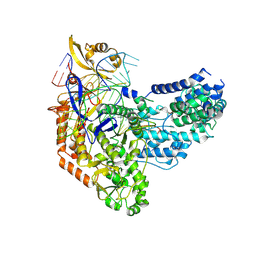 | | Cas9:sgRNA:DNA (S. pyogenes) with 0 RNA:DNA base pairs, open-protein/linear-DNA conformation | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1, Non-target DNA strand, Single-guide RNA, ... | | 著者 | Cofsky, J.C, Soczek, K.M, Knott, G.J, Nogales, E, Doudna, J.A. | | 登録日 | 2021-09-06 | | 公開日 | 2022-04-20 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | CRISPR-Cas9 bends and twists DNA to read its sequence.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7S38
 
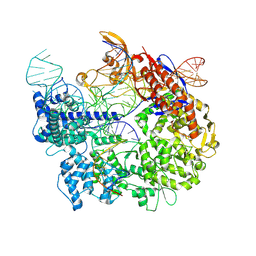 | | Cas9:sgRNA:DNA (S. pyogenes) forming a 3-base-pair R-loop | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1, Non-target DNA strand, Single-guide RNA, ... | | 著者 | Cofsky, J.C, Soczek, K.M, Knott, G.J, Nogales, E, Doudna, J.A. | | 登録日 | 2021-09-04 | | 公開日 | 2022-04-20 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | CRISPR-Cas9 bends and twists DNA to read its sequence.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5EAY
 
 | |
9CJH
 
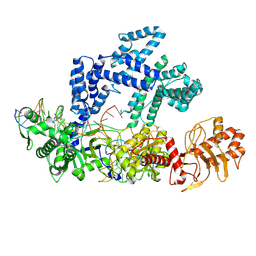 | |
9CJI
 
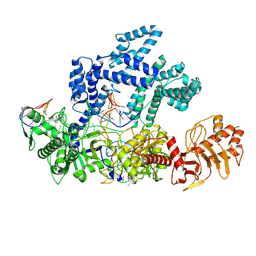 | |
9CJJ
 
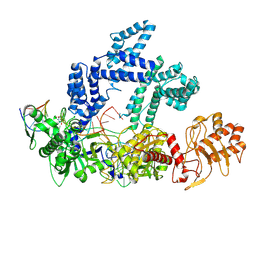 | |
4NRW
 
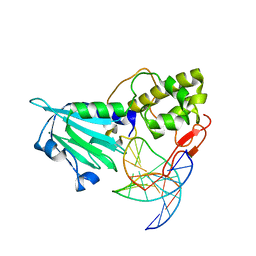 | | MvNei1-G86D | | 分子名称: | 5'-D(*CP*GP*TP*CP*CP*AP*(3DR)P*GP*TP*CP*TP*AP*C)-3', 5'-D(*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*G)-3', formamidopyrimidine-DNA glycosylase | | 著者 | Prakash, A, Doublie, S. | | 登録日 | 2013-11-27 | | 公開日 | 2014-01-01 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.845 Å) | | 主引用文献 | Genome and cancer single nucleotide polymorphisms of the human NEIL1 DNA glycosylase: Activity, structure, and the effect of editing.
Dna Repair, 14, 2014
|
|
8UZA
 
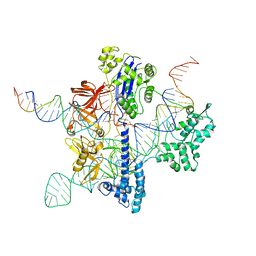 | | Cryo-EM structure of GeoCas9 in complex with sgRNA and target DNA | | 分子名称: | CRISPR-associated endonuclease Cas9, Non-target strand DNA, Target strand DNA, ... | | 著者 | Eggers, A.R, Soczek, K.M, Tuck, O.T, Doudna, J.A. | | 登録日 | 2023-11-14 | | 公開日 | 2024-05-29 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.17 Å) | | 主引用文献 | Rapid DNA unwinding accelerates genome editing by engineered CRISPR-Cas9.
Cell, 187, 2024
|
|
8UZB
 
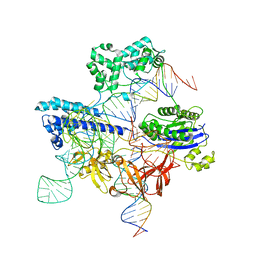 | | Cryo-EM structure of iGeoCas9 in complex with sgRNA and target DNA | | 分子名称: | CRISPR-associated endonuclease Cas9, Non-target strand DNA, RNA (107-MER), ... | | 著者 | Eggers, A.R, Soczek, K.M, Tuck, O.T, Doudna, J.A. | | 登録日 | 2023-11-14 | | 公開日 | 2024-05-29 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.63 Å) | | 主引用文献 | Rapid DNA unwinding accelerates genome editing by engineered CRISPR-Cas9.
Cell, 187, 2024
|
|
4MB7
 
 | |
7P0J
 
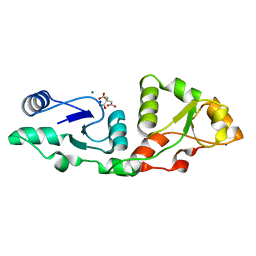 | | Crystal structure of S.pombe Mdb1 BRCT domains | | 分子名称: | CITRIC ACID, DNA damage response protein Mdb1, MAGNESIUM ION, ... | | 著者 | Day, M, Oliver, A.W, Pearl, L.H. | | 登録日 | 2021-06-29 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Phosphorylation-dependent assembly of DNA damage response systems and the central roles of TOPBP1.
DNA Repair (Amst), 108, 2021
|
|
4JGC
 
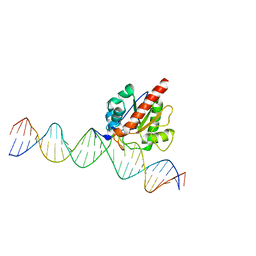 | | Human TDG N140A mutant IN A COMPLEX WITH 5-carboxylcytosine (5caC) | | 分子名称: | 4-amino-2-oxo-1,2-dihydropyrimidine-5-carboxylic acid, G/T mismatch-specific thymine DNA glycosylase, oligonucleotide, ... | | 著者 | Hashimoto, H, Zhang, X, Cheng, X. | | 登録日 | 2013-02-28 | | 公開日 | 2013-05-29 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.582 Å) | | 主引用文献 | Activity and crystal structure of human thymine DNA glycosylase mutant N140A with 5-carboxylcytosine DNA at low pH.
Dna Repair, 12, 2013
|
|
5F9R
 
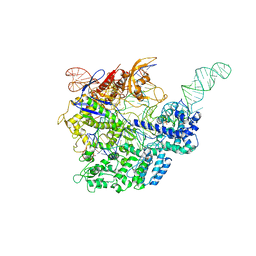 | |
5DS5
 
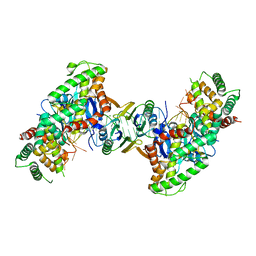 | | Crystal structure the Escherichia coli Cas1-Cas2 complex bound to protospacer DNA and Mg | | 分子名称: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (28-MER), ... | | 著者 | Nunez, J.K, Harrington, L.B, Kranzusch, P.J, Engelman, A.N, Doudna, J.A. | | 登録日 | 2015-09-16 | | 公開日 | 2015-10-28 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.951 Å) | | 主引用文献 | Foreign DNA capture during CRISPR-Cas adaptive immunity.
Nature, 527, 2015
|
|
5DS6
 
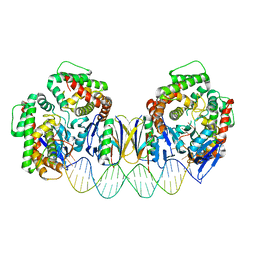 | | Crystal structure the Escherichia coli Cas1-Cas2 complex bound to protospacer DNA with splayed ends | | 分子名称: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (28-MER), ... | | 著者 | Nunez, J.K, Harrington, L.B, Kranzusch, P.J, Engelman, A.N, Doudna, J.A. | | 登録日 | 2015-09-16 | | 公開日 | 2015-10-28 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.352 Å) | | 主引用文献 | Foreign DNA capture during CRISPR-Cas adaptive immunity.
Nature, 527, 2015
|
|
5DS4
 
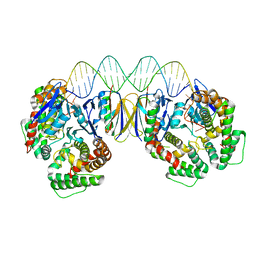 | | Crystal structure the Escherichia coli Cas1-Cas2 complex bound to protospacer DNA | | 分子名称: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (28-MER) | | 著者 | Nunez, J.K, Harrington, L.B, Kranzusch, P.J, Engelman, A.N, Doudna, J.A. | | 登録日 | 2015-09-16 | | 公開日 | 2015-10-28 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Foreign DNA capture during CRISPR-Cas adaptive immunity.
Nature, 527, 2015
|
|
6L6S
 
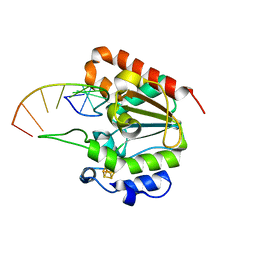 | |
7P8V
 
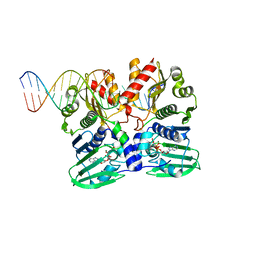 | |
4OU6
 
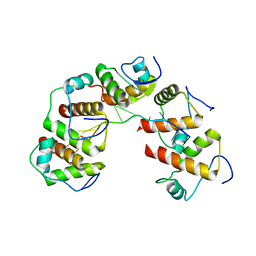 | | Crystal structure of DnaT84-153-dT10 ssDNA complex form 1 | | 分子名称: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), Primosomal protein 1 | | 著者 | Liu, Z, Chen, P, Niu, L, Teng, M, Li, X. | | 登録日 | 2014-02-15 | | 公開日 | 2014-08-13 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Crystal structure of DnaT84-153-dT10 ssDNA complex reveals a novel single-stranded DNA binding mode.
Nucleic Acids Res., 42, 2014
|
|
4OU7
 
 | | Crystal structure of DnaT84-153-dT10 ssDNA complex reveals a novel single-stranded DNA binding mode | | 分子名称: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), Primosomal protein 1 | | 著者 | Liu, Z, Chen, P, Niu, L, Teng, M, Li, X. | | 登録日 | 2014-02-15 | | 公開日 | 2014-08-13 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.83 Å) | | 主引用文献 | Crystal structure of DnaT84-153-dT10 ssDNA complex reveals a novel single-stranded DNA binding mode.
Nucleic Acids Res., 42, 2014
|
|
5EAX
 
 | | Crystal structure of Dna2 in complex with an ssDNA | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA replication ATP-dependent helicase/nuclease DNA2, ... | | 著者 | Zhou, C, Pourmal, S, Pavletich, N.P. | | 登録日 | 2015-10-17 | | 公開日 | 2015-11-18 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | Dna2 nuclease-helicase structure, mechanism and regulation by Rpa.
Elife, 4, 2015
|
|
5EAN
 
 | | Crystal structure of Dna2 in complex with a 5' overhang DNA | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, DNA (5'-D(P*AP*CP*TP*CP*TP*GP*CP*CP*AP*AP*GP*AP*GP*GP*A)-3'), ... | | 著者 | Zhou, C, Pourmal, S, Pavletich, N.P. | | 登録日 | 2015-10-16 | | 公開日 | 2015-11-18 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | Dna2 nuclease-helicase structure, mechanism and regulation by Rpa.
Elife, 4, 2015
|
|
5EAW
 
 | | Crystal structure of Dna2 nuclease-helicase | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA replication ATP-dependent helicase/nuclease DNA2, IRON/SULFUR CLUSTER | | 著者 | Zhou, C, Pourmal, S, Pavletich, N.P. | | 登録日 | 2015-10-17 | | 公開日 | 2015-11-18 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Dna2 nuclease-helicase structure, mechanism and regulation by Rpa.
Elife, 4, 2015
|
|
3R8F
 
 | | Replication initiator DnaA bound to AMPPCP and single-stranded DNA | | 分子名称: | 5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3', Chromosomal replication initiator protein dnaA, MAGNESIUM ION, ... | | 著者 | Duderstadt, K.E, Chuang, K, Berger, J.M. | | 登録日 | 2011-03-23 | | 公開日 | 2011-09-28 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3.366 Å) | | 主引用文献 | DNA stretching by bacterial initiators promotes replication origin opening.
Nature, 478, 2011
|
|
