8RGE
 
 | | Serial synchrotron in plate room temperature structure of Lysozyme. | | 分子名称: | CHLORIDE ION, Lysozyme C, SODIUM ION | | 著者 | Thompson, A.J, Hough, M.A, Sanchez-Weatherby, J, Williams, L.J, Sandy, J, Worrall, J.A.R. | | 登録日 | 2023-12-13 | | 公開日 | 2023-12-27 | | 最終更新日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Efficient in situ screening of and data collection from microcrystals in crystallization plates.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
6EG0
 
 | | Crystal structure of Dpr4 Ig1-Ig2 in complex with DIP-Eta Ig1-Ig3 | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Cosmanescu, F, Shapiro, L. | | 登録日 | 2018-08-17 | | 公開日 | 2018-11-28 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Neuron-Subtype-Specific Expression, Interaction Affinities, and Specificity Determinants of DIP/Dpr Cell Recognition Proteins.
Neuron, 100, 2018
|
|
6EFY
 
 | | Crystal Structure of DIP-Alpha Ig1-3 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Dpr-interacting protein alpha, isoform A, ... | | 著者 | Cosmanescu, F, Shapiro, L. | | 登録日 | 2018-08-17 | | 公開日 | 2018-11-28 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Neuron-Subtype-Specific Expression, Interaction Affinities, and Specificity Determinants of DIP/Dpr Cell Recognition Proteins.
Neuron, 100, 2018
|
|
6FF7
 
 | | human Bact spliceosome core structure | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-DIPHOSPHATE, BUD13 homolog, ... | | 著者 | Haselbach, D, Komarov, I, Agafonov, D, Hartmuth, K, Graf, B, Kastner, B, Luehrmann, R, Stark, H. | | 登録日 | 2018-01-03 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Structure and Conformational Dynamics of the Human Spliceosomal BactComplex.
Cell, 172, 2018
|
|
6C0F
 
 | | Yeast nucleolar pre-60S ribosomal subunit (state 2) | | 分子名称: | 5.8S rRNA, 60S ribosomal protein L13-A, 60S ribosomal protein L14-A, ... | | 著者 | Sanghai, Z.A, Miller, L, Barandun, J, Hunziker, M, Chaker-Margot, M, Klinge, S. | | 登録日 | 2017-12-29 | | 公開日 | 2018-03-14 | | 最終更新日 | 2020-01-08 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Modular assembly of the nucleolar pre-60S ribosomal subunit.
Nature, 556, 2018
|
|
4D0M
 
 | | Phosphatidylinositol 4-kinase III beta in a complex with Rab11a-GTP- gamma-S and the Rab-binding domain of FIP3 | | 分子名称: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, N-(5-(4-CHLORO-3-(2-HYDROXY-ETHYLSULFAMOYL)- PHENYLTHIAZOLE-2-YL)-ACETAMIDE, ... | | 著者 | Burke, J.E, Inglis, A.J, Perisic, O, Masson, G.R, McLaughlin, S.H, Rutaganira, F, Shokat, K.M, Williams, R.L. | | 登録日 | 2014-04-29 | | 公開日 | 2014-05-28 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (6 Å) | | 主引用文献 | Structures of Pi4Kiiibeta Complexes Show Simultaneous Recruitment of Rab11 and its Effectors.
Science, 344, 2014
|
|
6G05
 
 | |
6G07
 
 | |
6S01
 
 | | Structure of LEDGF PWWP domain bound H3K36 methylated nucleosome | | 分子名称: | Histone H2A, Histone H2B 1.1, Histone H3, ... | | 著者 | Wang, H, Farnung, L, Dienemann, C, Cramer, P. | | 登録日 | 2019-06-13 | | 公開日 | 2019-12-18 | | 最終更新日 | 2020-01-22 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structure of H3K36-methylated nucleosome-PWWP complex reveals multivalent cross-gyre binding.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6FZU
 
 | |
6RW4
 
 | | Structure of human mitochondrial 28S ribosome in complex with mitochondrial IF3 | | 分子名称: | 12S mitochondrial rRNA, 28S ribosomal protein S10, mitochondrial, ... | | 著者 | Itoh, Y, Khawaja, A, Rorbach, J, Amunts, A. | | 登録日 | 2019-06-03 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.97 Å) | | 主引用文献 | Distinct pre-initiation steps in human mitochondrial translation.
Nat Commun, 11, 2020
|
|
6RW5
 
 | | Structure of human mitochondrial 28S ribosome in complex with mitochondrial IF2 and IF3 | | 分子名称: | 12S mitochondrial rRNA, 28S ribosomal protein S10, mitochondrial, ... | | 著者 | Itoh, Y, Khawaja, A, Rorbach, J, Amunts, A. | | 登録日 | 2019-06-03 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.14 Å) | | 主引用文献 | Distinct pre-initiation steps in human mitochondrial translation.
Nat Commun, 11, 2020
|
|
3ZS2
 
 | | TyrB25,NMePheB26,LysB28,ProB29-insulin analogue crystal structure | | 分子名称: | CHLORIDE ION, INSULIN A CHAIN, INSULIN B CHAIN, ... | | 著者 | Antolikova, E, Zakova, L, Turkenburg, J.P, Watson, C.J, Hanclova, I, Sanda, M, Cooper, A, Kraus, T, Brzozowski, A.M, Jiracek, J.A. | | 登録日 | 2011-06-21 | | 公開日 | 2011-08-31 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Non-Equivalent Role of Inter- and Intramolecular Hydrogen Bonds in the Insulin Dimer Interface.
J.Biol.Chem., 286, 2011
|
|
1M7J
 
 | | Crystal structure of D-aminoacylase defines a novel subset of amidohydrolases | | 分子名称: | ACETATE ION, D-aminoacylase, ZINC ION | | 著者 | Liaw, S.-H, Chen, S.-J, Ko, T.-P, Hsu, C.-S, Wang, A.H.-J, Tsai, Y.-C. | | 登録日 | 2002-07-22 | | 公開日 | 2003-02-25 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal Structure of D-Aminoacylase from Alcaligenes faecalis DA1. A NOVEL SUBSET OF AMIDOHYDROLASES AND INSIGHTS INTO THE ENZYME MECHANISM.
J.Biol.Chem., 278, 2003
|
|
3ZQR
 
 | | NMePheB25 insulin analogue crystal structure | | 分子名称: | CHLORIDE ION, INSULIN A CHAIN, INSULIN B CHAIN, ... | | 著者 | Antolikova, E, Zakova, L, Turkenburg, J.P, Watson, C.J, Hanclova, I, Sanda, M, Cooper, A, Kraus, T, Brzozowski, A.M, Jiracek, J.A. | | 登録日 | 2011-06-10 | | 公開日 | 2011-08-24 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Non-Equivalent Role of Inter- and Intramolecular Hydrogen Bonds in the Insulin Dimer Interface.
J.Biol.Chem., 286, 2011
|
|
8WU8
 
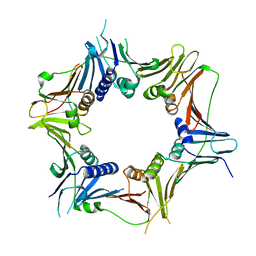 | | Crystal structure of the human RAD9-RAD1(F64A/M256A/F266A)-HUS1-RHINO(88-99) complex | | 分子名称: | Cell cycle checkpoint control protein RAD9A, Cell cycle checkpoint protein RAD1, Checkpoint protein HUS1, ... | | 著者 | Hara, K, Nagata, K, Iida, N, Hashimoto, H. | | 登録日 | 2023-10-20 | | 公開日 | 2024-02-14 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | Structural basis for intra- and intermolecular interactions on RAD9 subunit of 9-1-1 checkpoint clamp implies functional 9-1-1 regulation by RHINO.
J.Biol.Chem., 300, 2024
|
|
2Q5A
 
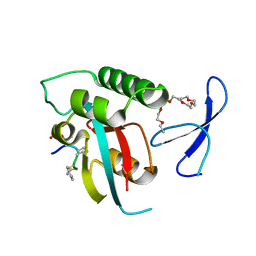 | | human Pin1 bound to L-PEPTIDE | | 分子名称: | 3,6,9,12,15,18-HEXAOXAICOSANE, Five residue peptide, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | 著者 | Noel, J.P, Zhang, Y. | | 登録日 | 2007-05-31 | | 公開日 | 2007-06-26 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural basis for high-affinity peptide inhibition of human Pin1.
Acs Chem.Biol., 2, 2007
|
|
5L1W
 
 | |
5L1V
 
 | |
4OYJ
 
 | |
5L1R
 
 | |
5L1U
 
 | |
5L1T
 
 | |
4KC8
 
 | | Crystal Structure of Endo-1,5-alpha-L-arabinanase from Thermotoga petrophila RKU-1 in complex with TRIS | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Glycoside hydrolase, ... | | 著者 | Nascimento, A.F.Z, Polo, C.C, Santos, C.R, Costa, M.C.M.F, Mesa, A.N, Prade, R.A, Ruller, R, Squina, F.M, Murakami, M.T. | | 登録日 | 2013-04-24 | | 公開日 | 2014-02-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Mechanistic strategies for catalysis adopted by evolutionary distinct family 43 arabinanases.
J.Biol.Chem., 289, 2014
|
|
4OYK
 
 | |
