6Q01
 
 | | TDP2 UBA Domain Bound to Ubiquitin at 0.85 Angstroms Resolution, Crystal Form 2 | | 分子名称: | 1,2-ETHANEDIOL, BENZOIC ACID, MAGNESIUM ION, ... | | 著者 | Schellenberg, M.J, Krahn, J.M, Williams, R.S. | | 登録日 | 2019-08-01 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (0.851 Å) | | 主引用文献 | Ubiquitin stimulated reversal of topoisomerase 2 DNA-protein crosslinks by TDP2.
Nucleic Acids Res., 48, 2020
|
|
6CIM
 
 | | Pre-Reaction Complex, RAG1(E962Q)/2-nicked/intact 12/23RSS complex in Mn2+ | | 分子名称: | DNA (5'-D(*GP*CP*CP*TP*GP*TP*CP*TP*TP*A)-3'), High mobility group protein B1, Intact 23RSS substrate forward strand, ... | | 著者 | Chuenchor, W, Chen, X, Kim, M.S, Gellert, M, Yang, W. | | 登録日 | 2018-02-24 | | 公開日 | 2018-04-25 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Cracking the DNA Code for V(D)J Recombination.
Mol. Cell, 70, 2018
|
|
5H1B
 
 | | Human RAD51 presynaptic complex | | 分子名称: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA repair protein RAD51 homolog 1, MAGNESIUM ION, ... | | 著者 | Xu, J, Zhao, L, Xu, Y, Zhao, W, Sung, P, Wang, H.W. | | 登録日 | 2016-10-08 | | 公開日 | 2016-12-21 | | 最終更新日 | 2022-03-23 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Cryo-EM structures of human RAD51 recombinase filaments during catalysis of DNA-strand exchange
Nat. Struct. Mol. Biol., 24, 2017
|
|
3VU7
 
 | | Crystal structure of REV1-REV7-REV3 ternary complex | | 分子名称: | DNA polymerase zeta catalytic subunit, DNA repair protein REV1, Mitotic spindle assembly checkpoint protein MAD2B | | 著者 | Kikuchi, S, Hara, K, Shimizu, T, Sato, M, Hashimoto, H. | | 登録日 | 2012-06-20 | | 公開日 | 2012-08-08 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural basis of recruitment of DNA polymerase [zeta] by interaction between REV1 and REV7 proteins
J.Biol.Chem., 287, 2012
|
|
5YDR
 
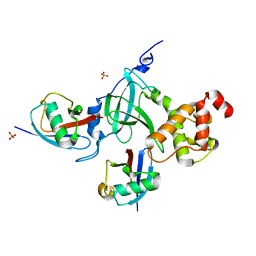 | | Structure of DNMT1 RFTS domain in complex with ubiquitin | | 分子名称: | DNA (cytosine-5)-methyltransferase 1, PHOSPHATE ION, Polyubiquitin-B, ... | | 著者 | Qian, C. | | 登録日 | 2017-09-14 | | 公開日 | 2018-02-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.003 Å) | | 主引用文献 | Structural and mechanistic insights into UHRF1-mediated DNMT1 activation in the maintenance DNA methylation.
Nucleic Acids Res., 46, 2018
|
|
6CIJ
 
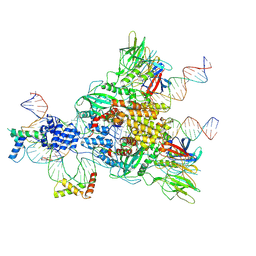 | | Cryo-EM structure of mouse RAG1/2 HFC complex containing partial HMGB1 linker(3.9 A) | | 分子名称: | CALCIUM ION, DNA (30-MER), DNA (41-MER), ... | | 著者 | Chen, X, Kim, M, Chuenchor, W, Cui, Y, Zhang, X, Zhou, Z.H, Gellert, M, Yang, W. | | 登録日 | 2018-02-24 | | 公開日 | 2018-04-25 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Cracking the DNA Code for V(D)J Recombination.
Mol. Cell, 70, 2018
|
|
1JEQ
 
 | |
7OPD
 
 | |
7OPC
 
 | |
7OO3
 
 | | Pol II-CSB-CSA-DDB1-UVSSA (Structure1) | | 分子名称: | CSB element, DNA damage-binding protein 1, DNA excision repair protein ERCC-6, ... | | 著者 | Kokic, G, Cramer, P. | | 登録日 | 2021-05-26 | | 公開日 | 2021-10-06 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structural basis of human transcription-DNA repair coupling.
Nature, 598, 2021
|
|
7OOB
 
 | |
3ZQS
 
 | | Human FANCL central domain | | 分子名称: | E3 UBIQUITIN-PROTEIN LIGASE FANCL, HEXAETHYLENE GLYCOL, PROLINE, ... | | 著者 | Hodson, C, Cole, A.R, Purkiss-Trew, A, Walden, H. | | 登録日 | 2011-06-10 | | 公開日 | 2011-07-20 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Analysis of Human Fancl, the E3 Ligase in the Fanconi Anemia Pathway.
J.Biol.Chem., 286, 2011
|
|
2IQC
 
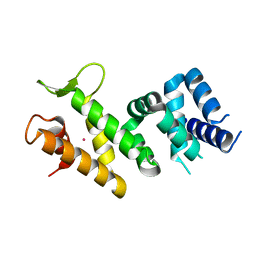 | | Crystal structure of Human FancF Protein that Functions in the Assembly of a DNA Damage Signaling Complex | | 分子名称: | Fanconi anemia group F protein, MERCURY (II) ION | | 著者 | Kowal, P, Gurtan, A.M, Stuckert, P, Lehmann, C, D'Andrea, A, Ellenberger, T.E. | | 登録日 | 2006-10-13 | | 公開日 | 2006-11-07 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural determinants of human FANCF protein that function in the assembly of a DNA damage signaling complex.
J.Biol.Chem., 282, 2007
|
|
7OKQ
 
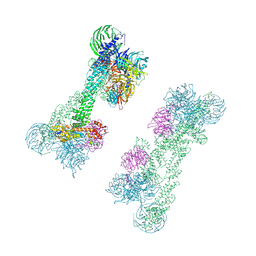 | | Cryo-EM Structure of the DDB1-DCAF1-CUL4A-RBX1 Complex | | 分子名称: | Cullin-4A, DDB1- and CUL4-associated factor 1, DNA damage-binding protein 1, ... | | 著者 | Mohamed, W.I, Schenk, A.D, Kempf, G, Cavadini, S, Thoma, N.H. | | 登録日 | 2021-05-18 | | 公開日 | 2021-10-13 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (8.4 Å) | | 主引用文献 | The CRL4 DCAF1 cullin-RING ubiquitin ligase is activated following a switch in oligomerization state.
Embo J., 40, 2021
|
|
8OHD
 
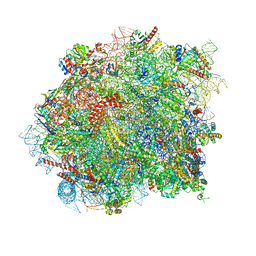 | | 60S ribosomal subunit bound to the E3-UFM1 complex - state 3 (native) | | 分子名称: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | 著者 | Penchev, I, DaRosa, P.A, Becker, T, Beckmann, R, Kopito, R. | | 登録日 | 2023-03-21 | | 公開日 | 2024-02-21 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | UFM1 E3 ligase promotes recycling of 60S ribosomal subunits from the ER.
Nature, 627, 2024
|
|
5HXB
 
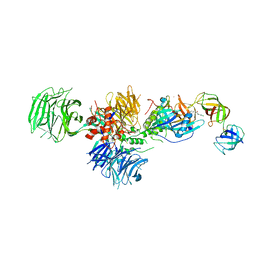 | | Cereblon in complex with DDB1, CC-885, and GSPT1 | | 分子名称: | 1-(3-chloro-4-methylphenyl)-3-({2-[(3S)-2,6-dioxopiperidin-3-yl]-1-oxo-2,3-dihydro-1H-isoindol-5-yl}methyl)urea, DNA damage-binding protein 1, Eukaryotic peptide chain release factor GTP-binding subunit ERF3A, ... | | 著者 | Chamberlain, P.P, Matyskiela, M, Pagarigan, B. | | 登録日 | 2016-01-30 | | 公開日 | 2016-06-29 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | A novel cereblon modulator recruits GSPT1 to the CRL4(CRBN) ubiquitin ligase.
Nature, 535, 2016
|
|
8OJ0
 
 | | 60S ribosomal subunit bound to the E3-UFM1 complex - state 2 (native) | | 分子名称: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | 著者 | Penchev, I, DaRosa, P.A, Becker, T, Beckmann, R, Kopito, R. | | 登録日 | 2023-03-23 | | 公開日 | 2024-02-21 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | UFM1 E3 ligase promotes recycling of 60S ribosomal subunits from the ER.
Nature, 627, 2024
|
|
8OJ5
 
 | | 60S ribosomal subunit bound to the E3-UFM1 complex - state 3 (in-vitro reconstitution) | | 分子名称: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | 著者 | Penchev, I, DaRosa, P.A, Peter, J.J, Kulathu, Y, Becker, T, Beckmann, R, Kopito, R. | | 登録日 | 2023-03-23 | | 公開日 | 2024-02-21 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | UFM1 E3 ligase promotes recycling of 60S ribosomal subunits from the ER.
Nature, 627, 2024
|
|
8W7M
 
 | | Yeast replisome in state V | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45, DNA (71-mer), ... | | 著者 | Dang, S, Zhai, Y, Feng, J, Yu, D, Xu, Z. | | 登録日 | 2023-08-30 | | 公開日 | 2023-12-06 | | 実験手法 | ELECTRON MICROSCOPY (4.12 Å) | | 主引用文献 | Synergism between CMG helicase and leading strand DNA polymerase at replication fork.
Nat Commun, 14, 2023
|
|
8W7S
 
 | | Yeast replisome in state IV | | 分子名称: | Cell division control protein 45, DNA (71-mer), DNA polymerase alpha-binding protein, ... | | 著者 | Dang, S, Zhai, Y, Feng, J, Yu, D. | | 登録日 | 2023-08-31 | | 公開日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (7.39 Å) | | 主引用文献 | Synergism between CMG helicase and leading strand DNA polymerase at replication fork.
Nat Commun, 14, 2023
|
|
4S3O
 
 | | PCGF5-RING1B-UbcH5c complex | | 分子名称: | E3 ubiquitin-protein ligase RING2, Polycomb group RING finger protein 5, Ubiquitin-conjugating enzyme E2 D3, ... | | 著者 | Taherbhoy, A.M, Cochran, A.G. | | 登録日 | 2015-03-23 | | 公開日 | 2015-07-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | BMI1-RING1B is an autoinhibited RING E3 ubiquitin ligase.
Nat Commun, 6, 2015
|
|
4A08
 
 | | Structure of hsDDB1-drDDB2 bound to a 13 bp CPD-duplex (purine at D-1 position) at 3.0 A resolution (CPD 1) | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-D(*AP*CP*GP*CP*GP*AP*(TTD)P*GP*CP*GP*CP*CP*C)-3', 5'-D(*TP*GP*GP*GP*CP*GP*CP*CP*CP*TP*CP*GP*CP*G)-3', ... | | 著者 | Scrima, A, Fischer, E.S, Iwai, S, Gut, H, Thoma, N.H. | | 登録日 | 2011-09-08 | | 公開日 | 2011-11-30 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation
Cell(Cambridge,Mass.), 147, 2011
|
|
8H3F
 
 | | Cryo-EM Structure of the KBTBD2-CRL3-CSN complex | | 分子名称: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | 著者 | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | 登録日 | 2022-10-08 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (6.73 Å) | | 主引用文献 | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H3A
 
 | | Cryo-EM Structure of the KBTBD2-CRL3~N8(removed)-CSN complex | | 分子名称: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | 著者 | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | 登録日 | 2022-10-08 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (7.51 Å) | | 主引用文献 | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H38
 
 | | Cryo-EM Structure of the KBTBD2-CRL3~N8-CSN(mutate) complex | | 分子名称: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | 著者 | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | 登録日 | 2022-10-08 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.25 Å) | | 主引用文献 | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
