8BCK
 
 | |
8BC5
 
 | |
8BBT
 
 | |
8BCL
 
 | |
7XRY
 
 | | Crystal structure of MERS main protease in complex with inhibitor YH-53 | | 分子名称: | N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide, ORF1a | | 著者 | Lin, C, Zhong, F.L, Zhou, X.L, Li, J, Zhang, J. | | 登録日 | 2022-05-12 | | 公開日 | 2022-12-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Structural Basis for the Inhibition of Coronaviral Main Proteases by a Benzothiazole-Based Inhibitor.
Viruses, 14, 2022
|
|
7XRS
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with inhibitor YH-53 | | 分子名称: | N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide, Replicase polyprotein 1a | | 著者 | Zhou, X.L, Zhong, F.L, Lin, C, Zeng, P, Zhang, J, Li, J. | | 登録日 | 2022-05-11 | | 公開日 | 2022-12-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Structural Basis for the Inhibition of Coronaviral Main Proteases by a Benzothiazole-Based Inhibitor.
Viruses, 14, 2022
|
|
7YF7
 
 | |
6Q96
 
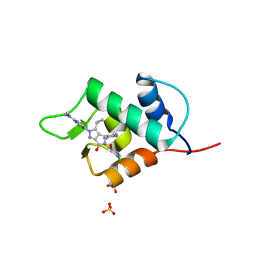 | |
7YGQ
 
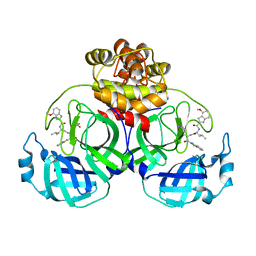 | | Crystal structure of SARS main protease in complex with inhibitor YH-53 | | 分子名称: | 3C-like proteinase nsp5, N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | 著者 | Lin, C, Zhong, F.L, Zhou, X.L, Zeng, P, Zhang, J, Li, J. | | 登録日 | 2022-07-12 | | 公開日 | 2022-12-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Structural Basis for the Inhibition of Coronaviral Main Proteases by a Benzothiazole-Based Inhibitor.
Viruses, 14, 2022
|
|
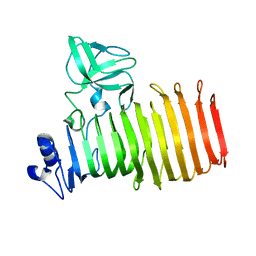
8BKE
 
 | |
8BMZ
 
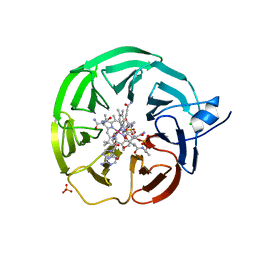 | | Bacteroides thetaiotaomicron surface lipoprotein BT1954 bound to adenosylcobalamin | | 分子名称: | Adenosylcobalamin, CHLORIDE ION, Putative surface layer protein, ... | | 著者 | Abellon-Ruiz, J, Jana, K, Silale, A, Basle, A, Kleinekathofer, U, van den Berg, B. | | 登録日 | 2022-11-11 | | 公開日 | 2023-08-16 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | BtuB TonB-dependent transporters and BtuG surface lipoproteins form stable complexes for vitamin B 12 uptake in gut Bacteroides.
Nat Commun, 14, 2023
|
|
7YCK
 
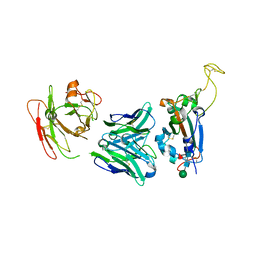 | | Crystal structure of SARS-CoV-2 Spike RBD in complex with FP-12A Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, FP-12A Fab heavy chain, FP-12A Fab light chain, ... | | 著者 | Nguyen, V.H.T, Chen, X. | | 登録日 | 2022-07-01 | | 公開日 | 2023-02-01 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis for a conserved neutralization epitope on the receptor-binding domain of SARS-CoV-2.
Nat Commun, 14, 2023
|
|
7YCL
 
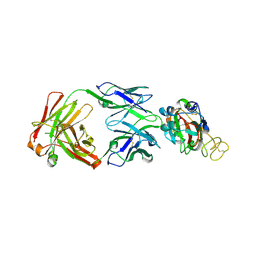 | | Crystal structure of SARS-CoV-2 Spike RBD in complex with IS-9A Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, IS-9A Fab heavy chain, IS-9A Fab light chain, ... | | 著者 | Mohapatra, A, Chen, X. | | 登録日 | 2022-07-01 | | 公開日 | 2023-02-01 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Structural basis for a conserved neutralization epitope on the receptor-binding domain of SARS-CoV-2.
Nat Commun, 14, 2023
|
|
7YCN
 
 | |
7WIV
 
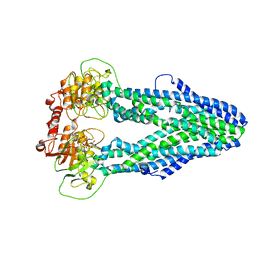 | | Cryo-EM structure of Mycobacterium tuberculosis irtAB in complex with an AMP-PNP | | 分子名称: | Mycobactin import ATP-binding/permease protein IrtA, Mycobactin import ATP-binding/permease protein IrtB, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Zhang, B, Sun, S, Yang, H, Rao, Z. | | 登録日 | 2022-01-05 | | 公開日 | 2023-01-25 | | 最終更新日 | 2024-04-10 | | 実験手法 | ELECTRON MICROSCOPY (2.88 Å) | | 主引用文献 | Cryo-EM structures for the Mycobacterium tuberculosis iron-loaded siderophore transporter IrtAB.
Protein Cell, 14, 2023
|
|
7WIU
 
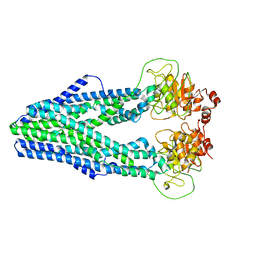 | | Cryo-EM structure of Mycobacterium tuberculosis irtAB in inward-facing state | | 分子名称: | Mycobactin import ATP-binding/permease protein IrtA, Mycobactin import ATP-binding/permease protein IrtB | | 著者 | Zhang, B, Sun, S, Yang, H, Rao, Z. | | 登録日 | 2022-01-05 | | 公開日 | 2023-01-25 | | 最終更新日 | 2024-04-10 | | 実験手法 | ELECTRON MICROSCOPY (3.48 Å) | | 主引用文献 | Cryo-EM structures for the Mycobacterium tuberculosis iron-loaded siderophore transporter IrtAB.
Protein Cell, 14, 2023
|
|
7WIW
 
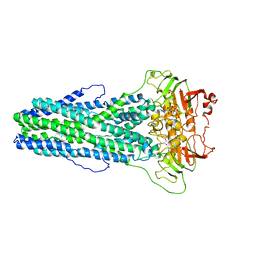 | | Cryo-EM structure of Mycobacterium tuberculosis irtAB complexed with ATP in an occluded conformation | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Mycobactin import ATP-binding/permease protein IrtA, ... | | 著者 | Zhang, B, Sun, S, Yang, H, Rao, Z. | | 登録日 | 2022-01-05 | | 公開日 | 2023-01-25 | | 最終更新日 | 2024-04-10 | | 実験手法 | ELECTRON MICROSCOPY (3.12 Å) | | 主引用文献 | Cryo-EM structures for the Mycobacterium tuberculosis iron-loaded siderophore transporter IrtAB.
Protein Cell, 14, 2023
|
|
8B8V
 
 | |
8BR9
 
 | |
8BJ7
 
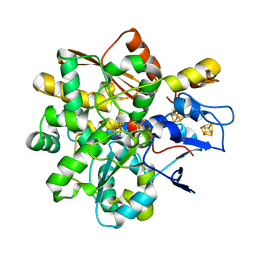 | |
7WN0
 
 | | Structure of PfENT1(Y190A) in complex with nanobody 19 | | 分子名称: | Equilibrative nucleoside/nucleobase transporter, nanobody19 | | 著者 | Wang, C, Deng, D, Ren, R.B, Yu, L.Y. | | 登録日 | 2022-01-17 | | 公開日 | 2023-02-01 | | 最終更新日 | 2023-08-16 | | 実験手法 | ELECTRON MICROSCOPY (3.64 Å) | | 主引用文献 | Structural basis of the substrate recognition and inhibition mechanism of Plasmodium falciparum nucleoside transporter PfENT1.
Nat Commun, 14, 2023
|
|
7WSX
 
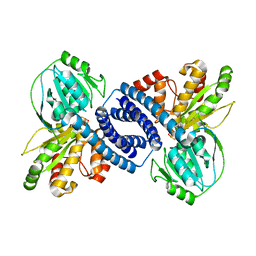 | |
7WN1
 
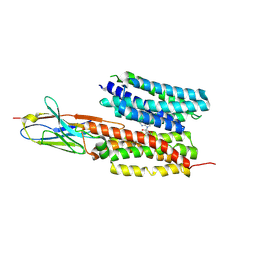 | | Structure of PfNT1(Y190A) in complex with nanobody 48 and inosine | | 分子名称: | Equilibrative nucleoside/nucleobase transporter, INOSINE, nanobody48 | | 著者 | Wang, C, Deng, D, Ren, R.B, Yu, L.Y. | | 登録日 | 2022-01-17 | | 公開日 | 2023-02-01 | | 最終更新日 | 2023-08-16 | | 実験手法 | ELECTRON MICROSCOPY (3.11 Å) | | 主引用文献 | Structural basis of the substrate recognition and inhibition mechanism of Plasmodium falciparum nucleoside transporter PfENT1.
Nat Commun, 14, 2023
|
|
7YAC
 
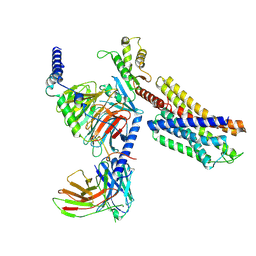 | | Paltusotine-bound SSTR2-Gi complex | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | 著者 | Zhao, J, Shao, Z. | | 登録日 | 2022-06-27 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-09-27 | | 実験手法 | ELECTRON MICROSCOPY (3.24 Å) | | 主引用文献 | Prospect of acromegaly therapy: molecular mechanism of clinical drugs octreotide and paltusotine.
Nat Commun, 14, 2023
|
|
7YIT
 
 | |
