3C3K
 
 | | Crystal structure of an uncharacterized protein from Actinobacillus succinogenes | | 分子名称: | Alanine racemase, CHLORIDE ION, GLYCEROL | | 著者 | Malashkevich, V.N, Toro, R, Meyer, A.J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2008-01-28 | | 公開日 | 2008-02-05 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Crystal structure of an uncharacterized protein from Actinobacillus succinogenes.
To be Published
|
|
6WWA
 
 | | Crystal structure of human SHLD2-SHLD3-REV7 complex | | 分子名称: | Mitotic spindle assembly checkpoint protein MAD2B, Shieldin complex subunit 2,Shieldin complex subunit 3 chimera | | 著者 | Xie, W, Patel, D.J. | | 登録日 | 2020-05-08 | | 公開日 | 2021-03-03 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Molecular mechanisms of assembly and TRIP13-mediated remodeling of the human Shieldin complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6WW9
 
 | |
3CB8
 
 | |
6WQS
 
 | | Xanthomonas citri Methionyl-tRNA synthetase in complex with REP8839 | | 分子名称: | 2-{[3-({[4-bromo-5-(1-fluoroethenyl)-3-methylthiophen-2-yl]methyl}amino)propyl]amino}quinolin-4(1H)-one, Methionine--tRNA ligase, ZINC ION | | 著者 | Mercaldi, G.F, Benedetti, C.E. | | 登録日 | 2020-04-29 | | 公開日 | 2021-04-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Molecular basis for diaryldiamine selectivity and competition with tRNA in a type 2 methionyl-tRNA synthetase from a Gram-negative bacterium.
J.Biol.Chem., 296, 2021
|
|
6WQT
 
 | | Xanthomonas citri Methionyl-tRNA synthetase in complex with REP3123 | | 分子名称: | 5-[(3-{[(4R)-6,8-dibromo-3,4-dihydro-2H-1-benzopyran-4-yl]amino}propyl)amino]thieno[3,2-b]pyridin-7(6H)-one, Methionine--tRNA ligase, ZINC ION | | 著者 | Mercaldi, G.F, Benedetti, C.E. | | 登録日 | 2020-04-29 | | 公開日 | 2021-04-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Molecular basis for diaryldiamine selectivity and competition with tRNA in a type 2 methionyl-tRNA synthetase from a Gram-negative bacterium.
J.Biol.Chem., 296, 2021
|
|
3GUR
 
 | | Crystal Structure of mu class glutathione S-transferase (GSTM2-2) in complex with glutathione and 6-(7-Nitro-2,1,3-benzoxadiazol-4-ylthio)hexanol (NBDHEX) | | 分子名称: | 1,2-ETHANEDIOL, GLUTATHIONE, Glutathione S-transferase Mu 2, ... | | 著者 | Federici, L, Lo Sterzo, C, Di Matteo, A, Scaloni, F, Federici, G, Caccuri, A.M. | | 登録日 | 2009-03-30 | | 公開日 | 2009-10-27 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural basis for the binding of the anticancer compound 6-(7-nitro-2,1,3-benzoxadiazol-4-ylthio)hexanol to human glutathione s-transferases
Cancer Res., 69, 2009
|
|
3ALN
 
 | | Crystal Structure of human non-phosphorylated MKK4 kinase domain complexed with AMP-PNP | | 分子名称: | Dual specificity mitogen-activated protein kinase kinase 4, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Matsumoto, T, Kinoshita, T, Kirii, Y, Yokota, K, Hamada, K, Tada, T. | | 登録日 | 2010-08-04 | | 公開日 | 2010-10-27 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structures of MKK4 kinase domain reveal that substrate peptide binds to an allosteric site and induces an auto-inhibition state
Biochem.Biophys.Res.Commun., 400, 2010
|
|
3CGO
 
 | | IRAK-4 Inhibitors (Part II)- A structure based assessment of imidazo[1,2 a]pyridine binding | | 分子名称: | 2-{4-[(4-imidazo[1,2-a]pyridin-3-ylpyrimidin-2-yl)amino]piperidin-1-yl}-N-methylacetamide, Mitogen-activated protein kinase 10 | | 著者 | Ceska, T.A, Platt, A, Fortunato, M, Dickson, K.M, Beevers, R. | | 登録日 | 2008-03-06 | | 公開日 | 2008-06-03 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | IRAK-4 inhibitors. Part II: A structure-based assessment of imidazo[1,2-a]pyridine binding
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3CGF
 
 | | IRAK-4 Inhibitors (Part II)- A structure based assessment of imidazo[1,2 a]pyridine binding | | 分子名称: | Mitogen-activated protein kinase 10, N-cyclohexyl-4-imidazo[1,2-a]pyridin-3-yl-N-methylpyrimidin-2-amine | | 著者 | Ceska, T.A, Platt, A, Fortunato, M, Dickson, K.M, Buckley, G. | | 登録日 | 2008-03-05 | | 公開日 | 2008-06-03 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | IRAK-4 inhibitors. Part II: A structure-based assessment of imidazo[1,2-a]pyridine binding
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3DA6
 
 | | Crystal Structure of human JNK3 complexed with N-(3-methyl-4-(3-(2-(methylamino)pyrimidin-4-yl)pyridin-2-yloxy)naphthalen-1-yl)-1H-benzo[d]imidazol-2-amine | | 分子名称: | Mitogen-activated protein kinase 10, N-[3-methyl-4-({3-[2-(methylamino)pyrimidin-4-yl]pyridin-2-yl}oxy)naphthalen-1-yl]-1H-benzimidazol-2-amine | | 著者 | Cee, V.J, Cheng, A.C, Romero, K, Bellon, S, Mohr, C, Whittington, D.A, Bready, J, Caenepeel, S, Coxon, A, Deak, H.L, Hodous, B.L, Kim, J.L, Lin, J, Nguyen, H, Olivieri, P.R, Patel, V.F, Wang, L, Hughes, P, Geuns-Meyer, S. | | 登録日 | 2008-05-28 | | 公開日 | 2009-01-06 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Pyridyl-pyrimidine benzimidazole derivatives as potent, selective, and orally bioavailable inhibitors of Tie-2 kinase
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3CES
 
 | | Crystal Structure of E.coli MnmG (GidA), a Highly-Conserved tRNA Modifying Enzyme | | 分子名称: | tRNA uridine 5-carboxymethylaminomethyl modification enzyme gidA | | 著者 | Shi, R, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | 登録日 | 2008-02-29 | | 公開日 | 2009-03-03 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.412 Å) | | 主引用文献 | Structure-function analysis of Escherichia coli MnmG (GidA), a highly conserved tRNA-modifying enzyme.
J.Bacteriol., 191, 2009
|
|
7VP9
 
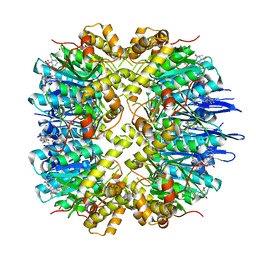 | | Crystal structure of human ClpP in complex with ZG111 | | 分子名称: | (6S,9aS)-N-[(4-bromophenyl)methyl]-6-[(2S)-butan-2-yl]-8-(naphthalen-1-ylmethyl)-4,7-bis(oxidanylidene)-3,6,9,9a-tetrahydro-2H-pyrazino[1,2-a]pyrimidine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit, mitochondrial, ... | | 著者 | Wang, P.Y, Gan, J.H, Yang, C.-G. | | 登録日 | 2021-10-15 | | 公開日 | 2022-06-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.552 Å) | | 主引用文献 | Aberrant human ClpP activation disturbs mitochondrial proteome homeostasis to suppress pancreatic ductal adenocarcinoma.
Cell Chem Biol, 29, 2022
|
|
7VPR
 
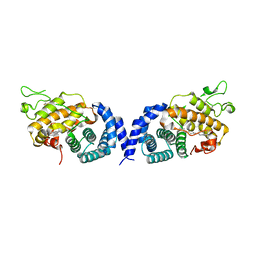 | |
7VPU
 
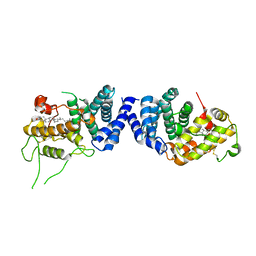 | |
7VPS
 
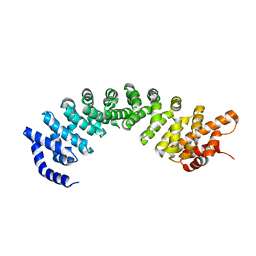 | |
7VPT
 
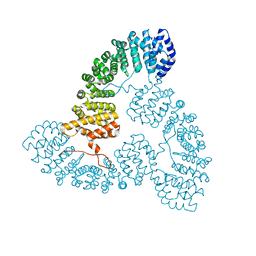 | |
7WKJ
 
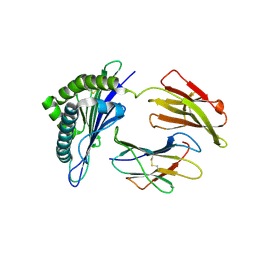 | | A COVID-19 T-cell response detection method based on a newly identified human CD8+ T cell epitope from SARS-CoV-2-Hubei Province, 2021. | | 分子名称: | Beta-2-microglobulin, LYS-THR-PHE-PRO-PRO-THR-GLU-PRO-LYS, MHC class I antigen | | 著者 | Zhang, J, Lu, D, Li, M, Liu, M.S, Yao, S.J, Zhan, J.B, Liu, J, Gao, G.F. | | 登録日 | 2022-01-10 | | 公開日 | 2022-03-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | A COVID-19 T-Cell Response Detection Method Based on a Newly Identified Human CD8 + T Cell Epitope from SARS-CoV-2 - Hubei Province, China, 2021.
China CDC Wkly, 4, 2022
|
|
7XJT
 
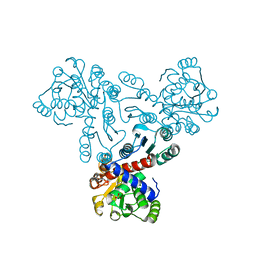 | |
7X99
 
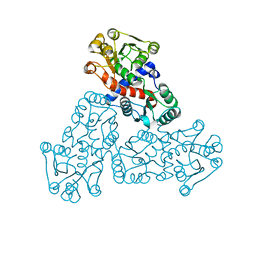 | |
7XB5
 
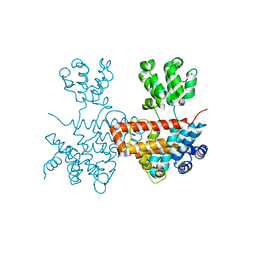 | |
7WWG
 
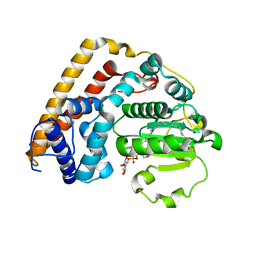 | | Crystal structure of Saccharomyces cerevisiae Sfh2 complexed with phosphatidylinositol in an open conformation | | 分子名称: | (1R)-2-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Phosphatidylinositol transfer protein CSR1 | | 著者 | Chen, L, Tan, L, Im, Y.J. | | 登録日 | 2022-02-12 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Structural basis of ligand recognition and transport by Sfh2, a yeast phosphatidylinositol transfer protein of the Sec14 superfamily.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7WWE
 
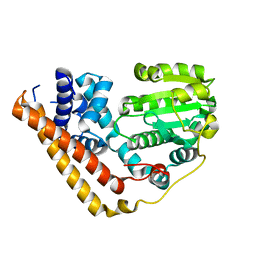 | |
7WWD
 
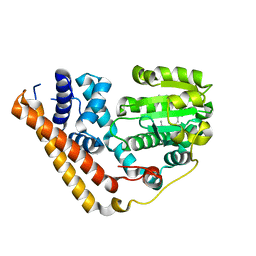 | | Crystal structure of Saccharomyces cerevisiae Sfh2 complexed with squalene | | 分子名称: | (6E,10E,14E,18E)-2,6,10,15,19,23-hexamethyltetracosa-2,6,10,14,18,22-hexaene, Phosphatidylinositol transfer protein CSR1 | | 著者 | Chen, L, Tan, L, Im, Y.J. | | 登録日 | 2022-02-12 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Structural basis of ligand recognition and transport by Sfh2, a yeast phosphatidylinositol transfer protein of the Sec14 superfamily.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7WVT
 
 | | Crystal structure of Saccharomyces cerevisiae Sfh2 complexed with phosphatidylinositol | | 分子名称: | (1R)-2-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Phosphatidylinositol transfer protein CSR1 | | 著者 | Chen, L, Tan, L, Im, Y.J. | | 登録日 | 2022-02-11 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural basis of ligand recognition and transport by Sfh2, a yeast phosphatidylinositol transfer protein of the Sec14 superfamily.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
