5JIK
 
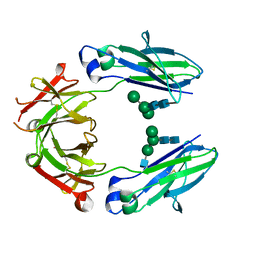 | | Crystal structure of HER2 binding IgG1-Fc (Fcab H10-03-6) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-1 chain C region, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Humm, A, Lobner, E, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | 登録日 | 2016-04-22 | | 公開日 | 2017-04-12 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Fcab-HER2 Interaction: a Menage a Trois. Lessons from X-Ray and Solution Studies.
Structure, 25, 2017
|
|
3ZOX
 
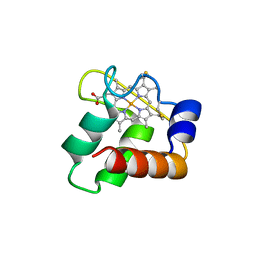 | | Crystal Structure of N64Del Mutant of Nitrosomonas europaea Cytochrome c552 (monoclinic space group) | | 分子名称: | CYTOCHROME C-552, HEME C | | 著者 | Hersleth, H.-P, Can, M, Krucinska, J, Zoppellaro, G, Andersen, N.H, Wedekind, J.E, Andersson, K.K, Bren, K.L. | | 登録日 | 2013-02-26 | | 公開日 | 2013-08-14 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural Characterization of Nitrosomonas Europaea Cytochrome C-552 Variants with Marked Differences in Electronic Structure.
Chembiochem, 14, 2013
|
|
5Z3N
 
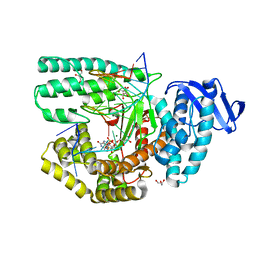 | | Structure of large fragment of DNA Polymerase I from Thermus aquaticus Host-Guest complex with the unnatural base 5fC pair with dA | | 分子名称: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*CP*GP*GP*CP*GP*CP*CP*GP*(5FC)P*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), ... | | 著者 | Zeng, H, Mondal, M, Song, R.Y, Zhang, J, Xia, B, Gao, Y.Q, Yi, C.Q. | | 登録日 | 2018-01-08 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Unnatural Cytosine Bases Recognized as Thymines by DNA Polymerases by the Formation of the Watson-Crick Geometry.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
1A1Q
 
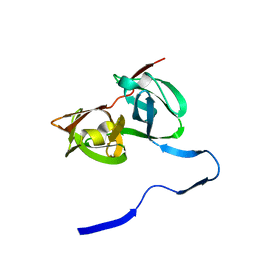 | | HEPATITIS C VIRUS NS3 PROTEINASE | | 分子名称: | NS3 PROTEINASE, ZINC ION | | 著者 | Love, R.A, Parge, H.E, Wickersham, J.A, Hostomsky, Z, Habuka, N, Moomaw, E.W, Adachi, T, Hostomska, Z. | | 登録日 | 1997-12-12 | | 公開日 | 1998-03-25 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The crystal structure of hepatitis C virus NS3 proteinase reveals a trypsin-like fold and a structural zinc binding site.
Cell(Cambridge,Mass.), 87, 1996
|
|
149D
 
 | |
6UL3
 
 | |
7Z3J
 
 | | Structure of crystallisable rat Phospholipase C gamma 1 in complex with inositol 1,4,5-trisphosphate | | 分子名称: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-1, CALCIUM ION, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, ... | | 著者 | Pinotsis, N, Bunney, T.D, Katan, M. | | 登録日 | 2022-03-02 | | 公開日 | 2022-07-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Characterization of the membrane interactions of phospholipase C gamma reveals key features of the active enzyme.
Sci Adv, 8, 2022
|
|
6UKC
 
 | |
1W1W
 
 | | Sc Smc1hd:Scc1-C complex, ATPgS | | 分子名称: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, SISTER CHROMATID COHESION PROTEIN 1, ... | | 著者 | Haering, C, Nasmyth, K, Lowe, J. | | 登録日 | 2004-06-24 | | 公開日 | 2004-09-30 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structure and stability of cohesin's Smc1-kleisin interaction.
Mol. Cell, 15, 2004
|
|
5NPI
 
 | |
6S0B
 
 | | Crystal Structure of Properdin in complex with the CTC domain of C3/C3b | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C3, Properdin, ... | | 著者 | van den Bos, R.M, Pearce, N.M, Gros, P. | | 登録日 | 2019-06-14 | | 公開日 | 2019-09-04 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.312 Å) | | 主引用文献 | Insights Into Enhanced Complement Activation by Structures of Properdin and Its Complex With the C-Terminal Domain of C3b.
Front Immunol, 10, 2019
|
|
6UE3
 
 | |
6S0A
 
 | | Crystal Structure of Properdin (TSR domains N12 & 456) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Properdin, alpha-D-mannopyranose, ... | | 著者 | van den Bos, R.M, Pearce, N.M, Gros, P. | | 登録日 | 2019-06-14 | | 公開日 | 2019-09-04 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.52 Å) | | 主引用文献 | Insights Into Enhanced Complement Activation by Structures of Properdin and Its Complex With the C-Terminal Domain of C3b.
Front Immunol, 10, 2019
|
|
7Y5B
 
 | | Cryo-EM structure of F-ATP synthase from Mycolicibacterium smegmatis (rotational state 1) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | 著者 | Saw, W.-G, Wong, C.F, Grueber, G. | | 登録日 | 2022-06-16 | | 公開日 | 2022-11-23 | | 最終更新日 | 2023-01-11 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Structural Elements Involved in ATP Hydrolysis Inhibition and ATP Synthesis of Tuberculosis and Nontuberculous Mycobacterial F-ATP Synthase Decipher New Targets for Inhibitors.
Antimicrob.Agents Chemother., 66, 2022
|
|
5YZE
 
 | | Crystal structure of the [Co2+-(chromomycin A3)2]-d(CCG)3 complex | | 分子名称: | (1S)-5-deoxy-1-O-methyl-1-C-[(2R,3S)-3,5,7,10-tetrahydroxy-6-methyl-4-oxo-1,2,3,4-tetrahydroanthracen-2-yl]-D-xylulose, 2,6-dideoxy-4-O-methyl-alpha-D-galactopyranose-(1-3)-(2R,3R,6R)-6-hydroxy-2-methyltetrahydro-2H-pyran-3-yl acetate, 3-C-methyl-4-O-acetyl-alpha-L-Olivopyranose-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol, ... | | 著者 | Hou, M.H, Chen, Y.W, Wu, P.C, Satange, R.B. | | 登録日 | 2017-12-14 | | 公開日 | 2018-10-17 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | CoII(Chromomycin)2 Complex Induces a Conformational Change of CCG Repeats from i-Motif to Base-Extruded DNA Duplex
Int J Mol Sci, 19, 2018
|
|
6IJX
 
 | | Crystal Structure of AKR1C1 complexed with meclofenamic acid | | 分子名称: | 2-[(2,6-dichloro-3-methyl-phenyl)amino]benzoic acid, Aldo-keto reductase family 1 member C1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Zheng, X, Zhao, Y, Zhang, L, Zhang, H, Chen, Y, Hu, X. | | 登録日 | 2018-10-12 | | 公開日 | 2019-10-16 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Screening, synthesis, crystal structure, and molecular basis of 6-amino-4-phenyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitriles as novel AKR1C3 inhibitors.
Bioorg.Med.Chem., 26, 2018
|
|
5LSI
 
 | | CRYSTAL STRUCTURE OF THE KINETOCHORE MIS12 COMPLEX HEAD2 SUBDOMAIN CONTAINING DSN1 AND NSL1 FRAGMENTS | | 分子名称: | Kinetochore-associated protein DSN1 homolog, Kinetochore-associated protein NSL1 homolog, SULFATE ION | | 著者 | Vetter, I.R, Petrovic, A, Keller, J, Liu, Y. | | 登録日 | 2016-09-02 | | 公開日 | 2016-11-16 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.002 Å) | | 主引用文献 | Structure of the MIS12 Complex and Molecular Basis of Its Interaction with CENP-C at Human Kinetochores.
Cell, 167, 2016
|
|
6W98
 
 | | Single-Particle Cryo-EM Structure of Arabinofuranosyltransferase AftD from Mycobacteria | | 分子名称: | 4'-PHOSPHOPANTETHEINE, Acyl carrier protein, CALCIUM ION, ... | | 著者 | Tan, Y.Z, Zhang, L, Rodrigues, J, Zheng, R.B, Giacometti, S.I, Rosario, A.L, Kloss, B, Dandey, V.P, Wei, H, Brunton, R, Raczkowski, A.M, Athayde, D, Catalao, M.J, Pimentel, M, Clarke, O.B, Lowary, T.L, Archer, M, Niederweis, M, Potter, C.S, Carragher, B, Mancia, F. | | 登録日 | 2020-03-22 | | 公開日 | 2020-05-13 | | 最終更新日 | 2020-06-03 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Cryo-EM Structures and Regulation of Arabinofuranosyltransferase AftD from Mycobacteria.
Mol.Cell, 78, 2020
|
|
4A3V
 
 | | yeast regulatory particle proteasome assembly chaperone Hsm3 in complex with Rpt1 C-terminal fragment | | 分子名称: | 26S PROTEASE REGULATORY SUBUNIT 7 HOMOLOG, DNA MISMATCH REPAIR PROTEIN HSM3, LINKER | | 著者 | Richet, N, Barrault, M.B, Godart, C, Murciano, B, Le Tallec, B, Rousseau, E, Ledu, M.H, Charbonnier, J.B, Legrand, P, Guerois, R, Peyroche, A, Ochsenbein, F. | | 登録日 | 2011-10-04 | | 公開日 | 2012-04-11 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Dual Functions of the Hsm3 Protein in Chaperoning and Scaffolding Regulatory Particle Subunits During the Proteasome Assembly.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8IUG
 
 | | Cryo-EM structure of the RC-LH core complex from roseiflexus castenholzii | | 分子名称: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 2-O-octyl-beta-D-glucopyranose, ... | | 著者 | Wang, G.-L, Qi, C.-H, Yu, L.-J. | | 登録日 | 2023-03-24 | | 公開日 | 2023-11-22 | | 最終更新日 | 2023-12-06 | | 実験手法 | ELECTRON MICROSCOPY (2.86 Å) | | 主引用文献 | New insights on the photocomplex of Roseiflexus castenholzii revealed from comparisons of native and carotenoid-depleted complexes.
J.Biol.Chem., 299, 2023
|
|
8IUN
 
 | | Cryo-EM structure of the CRT-LESS RC-LH core complex from roseiflexus castenholzii | | 分子名称: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 2-O-octyl-beta-D-glucopyranose, ... | | 著者 | Wang, G.-L, Qi, C.-H, Yu, L.-J. | | 登録日 | 2023-03-24 | | 公開日 | 2023-12-06 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | New insights on the photocomplex of Roseiflexus castenholzii revealed from comparisons of native and carotenoid-depleted complexes.
J.Biol.Chem., 299, 2023
|
|
5V6N
 
 | |
7XWN
 
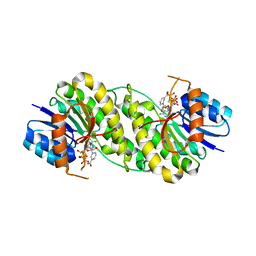 | | structure of patulin-detoxifying enzyme Y155F/V187K with NADPH and substrate | | 分子名称: | (4~{S})-4-oxidanyl-4,6-dihydrofuro[3,2-c]pyran-2-one, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase | | 著者 | Dai, L, Li, H, Hu, Y, Guo, R.T, Chen, C.C. | | 登録日 | 2022-05-26 | | 公開日 | 2022-10-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure-based rational design of a short-chain dehydrogenase/reductase for improving activity toward mycotoxin patulin.
Int.J.Biol.Macromol., 222, 2022
|
|
7XWK
 
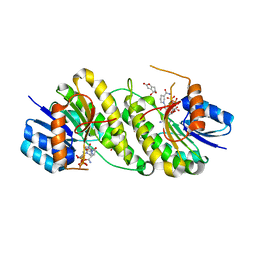 | | structure of patulin-detoxifying enzyme Y155F with NADPH and substrate | | 分子名称: | (4~{S})-4-oxidanyl-4,6-dihydrofuro[3,2-c]pyran-2-one, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase | | 著者 | Dai, L, Li, H, Hu, Y, Guo, R.T, Chen, C.C. | | 登録日 | 2022-05-26 | | 公開日 | 2022-10-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Structure-based rational design of a short-chain dehydrogenase/reductase for improving activity toward mycotoxin patulin.
Int.J.Biol.Macromol., 222, 2022
|
|
5UVW
 
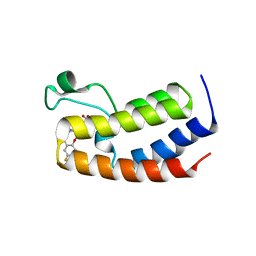 | | BRD4_Bromodomain1-A1376855 | | 分子名称: | Bromodomain-containing protein 4, N-[4-(2,4-difluorophenoxy)-3-(6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-4-yl)phenyl]ethanesulfonamide, SULFATE ION | | 著者 | Park, C.H. | | 登録日 | 2017-02-20 | | 公開日 | 2017-06-21 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | BRD4_Bromodomain1-A1376855
To Be Published
|
|
