5LO2
 
 | | Engineering protein stability with atomic precision in a monomeric miniprotein | | 分子名称: | PPaTyr | | 著者 | Baker, E.G, Hudson, K.L, Williams, C, Bartlett, G.G, Heal, J.W, Sessions, R.B, Crump, M.P, Woolfson, D.N. | | 登録日 | 2016-08-08 | | 公開日 | 2017-05-17 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Engineering protein stability with atomic precision in a monomeric miniprotein.
Nat. Chem. Biol., 13, 2017
|
|
5LO3
 
 | | Engineering protein stability with atomic precision in a monomeric miniprotein | | 分子名称: | PPaOMe | | 著者 | Baker, E.G, Hudson, K.L, Williams, C, Bartlett, G.G, Heal, J.W, Sessions, R.B, Crump, M.P, Woolfson, D.N. | | 登録日 | 2016-08-08 | | 公開日 | 2017-05-17 | | 最終更新日 | 2023-11-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Engineering protein stability with atomic precision in a monomeric miniprotein.
Nat. Chem. Biol., 13, 2017
|
|
6P78
 
 | | queuine lyase from Clostridium spiroforme bound to SAM and queuine | | 分子名称: | 2-amino-5-({[(1S,4S,5S)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-1,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, IRON/SULFUR CLUSTER, Queuine lyase, ... | | 著者 | Almo, S.C, Grove, T.L. | | 登録日 | 2019-06-05 | | 公開日 | 2019-09-18 | | 最終更新日 | 2019-12-18 | | 実験手法 | X-RAY DIFFRACTION (1.726 Å) | | 主引用文献 | Discovery of novel bacterial queuine salvage enzymes and pathways in human pathogens.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1I9F
 
 | | STRUCTURAL CHARACTERIZATION OF THE COMPLEX OF THE REV RESPONSE ELEMENT RNA WITH A SELECTED PEPTIDE | | 分子名称: | REV RESPONSE ELEMENT RNA, RSG-1.2 PEPTIDE | | 著者 | Zhang, Q, Harada, K, Cho, H.S, Frankel, A, Wemmer, D.E. | | 登録日 | 2001-03-19 | | 公開日 | 2001-05-25 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural characterization of the complex of the Rev response element RNA with a selected peptide.
Chem.Biol., 8, 2001
|
|
6CFE
 
 | |
8B6N
 
 | |
8B6P
 
 | |
6QAM
 
 | |
1GX5
 
 | |
5EMS
 
 | | Crystal Structure of an iodinated insulin analog | | 分子名称: | CHLORIDE ION, Insulin, PHENOL, ... | | 著者 | Lawrence, M.C, Pandyarajan, V, Wan, Z, Weiss, M.A. | | 登録日 | 2015-11-06 | | 公開日 | 2016-11-16 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Extending Halogen-based Medicinal Chemistry to Proteins: IODO-INSULIN AS A CASE STUDY.
J. Biol. Chem., 291, 2016
|
|
1GX6
 
 | |
2XWH
 
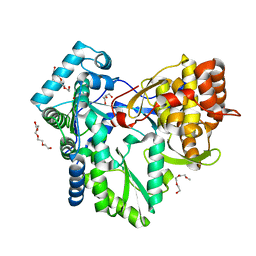 | | HCV-J6 NS5B polymerase structure at 1.8 Angstrom | | 分子名称: | DI(HYDROXYETHYL)ETHER, POLYETHYLENE GLYCOL (N=34), RNA DEPENDENT RNA POLYMERASE | | 著者 | Scrima, N, Bressanelli, S. | | 登録日 | 2010-11-03 | | 公開日 | 2011-01-12 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | A Comprehensive Structure-Function Comparison of Hepatitis C Virus Strains Jfh1 and J6 Polymerases Reveals a Key Residue Stimulating Replication in Cell Culture Across Genotypes.
J.Virol., 85, 2011
|
|
7YH8
 
 | | Crystal structure of a heterochiral protein complex | | 分子名称: | D-Pep-1, L-19437 | | 著者 | Liang, M, Li, S, Wang, T, Liu, L, Lu, P. | | 登録日 | 2022-07-13 | | 公開日 | 2023-07-19 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of a heterochiral protein complex
To Be Published
|
|
8TW0
 
 | | Crystal Structure of a synthetic ABC heterotrimeric Collagen-like Peptide at 1.53 A | | 分子名称: | Collagen Mimetic Peptide A, Collagen Mimetic Peptide B, Collagen Mimetic Peptide C, ... | | 著者 | Miller, M.D, Cole, C.C, Xu, W, Walker, D.R, Hulgan, S.A.H, Pogostin, B.H, Swain, J.W.R, Duella, R, Misiura, M, Wang, X, Kolomeisky, A.B, Phillips Jr, G.N, Hartgerink, J.D. | | 登録日 | 2023-08-18 | | 公開日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | Heterotrimeric Collagen Helix with High Specificity of Assembly Results in a Rapid Rate of Folding
Nat.Chem., 2024
|
|
8TQM
 
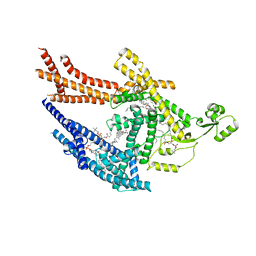 | |
8UBZ
 
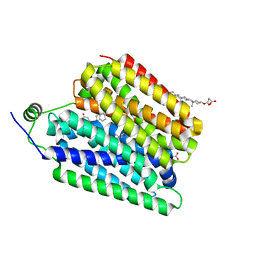 | | Choline-bound FLVCR1 | | 分子名称: | CHOLESTEROL HEMISUCCINATE, CHOLINE ION, Heme transporter FLVCR1 | | 著者 | Hite, R.K, Son, Y. | | 登録日 | 2023-09-25 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.02 Å) | | 主引用文献 | Structural basis of lipid head group entry to the Kennedy pathway by FLVCR1.
Nature, 629, 2024
|
|
8TQS
 
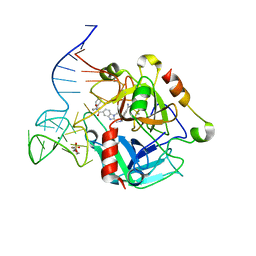 | |
8UBX
 
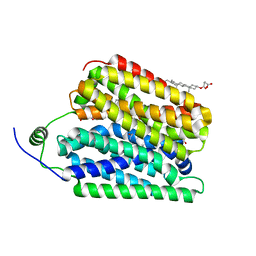 | | Ethanolamine-bound FLVCR1 | | 分子名称: | CHOLESTEROL HEMISUCCINATE, ETHANOLAMINE, Heme transporter FLVCR1 | | 著者 | Hite, R.K, Son, Y. | | 登録日 | 2023-09-25 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Structural basis of lipid head group entry to the Kennedy pathway by FLVCR1.
Nature, 629, 2024
|
|
8WOW
 
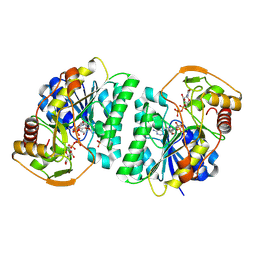 | | Crystal structure of Arabidopsis thaliana UDP-glucose 4-epimerase 2 (AtUGE2) complexed with UDP, I160L/G233A mutant | | 分子名称: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, UDP-glucose 4-epimerase 2, ... | | 著者 | Matsumoto, M, Umezawa, A, Kotake, T, Fushinobu, S. | | 登録日 | 2023-10-08 | | 公開日 | 2024-05-15 | | 最終更新日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Cytosolic UDP-L-arabinose synthesis by bifunctional UDP-glucose 4-epimerases in Arabidopsis.
Plant J., 119, 2024
|
|
8UBY
 
 | | Choline-bound FLVCR1 | | 分子名称: | CHOLESTEROL HEMISUCCINATE, CHOLINE ION, Heme transporter FLVCR1 | | 著者 | Hite, R.K, Son, Y. | | 登録日 | 2023-09-25 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.67 Å) | | 主引用文献 | Structural basis of lipid head group entry to the Kennedy pathway by FLVCR1.
Nature, 629, 2024
|
|
8UC0
 
 | | Endogenous ligand bound FLVCR1 | | 分子名称: | CHOLESTEROL HEMISUCCINATE, Heme transporter FLVCR1 | | 著者 | Hite, R.K, Son, Y. | | 登録日 | 2023-09-25 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.42 Å) | | 主引用文献 | Structural basis of lipid head group entry to the Kennedy pathway by FLVCR1.
Nature, 629, 2024
|
|
8WOV
 
 | | Crystal structure of Arabidopsis thaliana UDP-glucose 4-epimerase 2 (AtUGE2) complexed with UDP, G233A mutant | | 分子名称: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase 2, URIDINE-5'-DIPHOSPHATE | | 著者 | Matsumoto, M, Umezawa, A, Kotake, T, Fushinobu, S. | | 登録日 | 2023-10-07 | | 公開日 | 2024-05-15 | | 最終更新日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Cytosolic UDP-L-arabinose synthesis by bifunctional UDP-glucose 4-epimerases in Arabidopsis.
Plant J., 119, 2024
|
|
8WOP
 
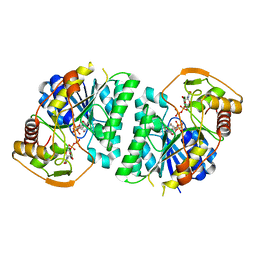 | | Crystal structure of Arabidopsis thaliana UDP-glucose 4-epimerase 2 (AtUGE2) complexed with UDP, wild-type | | 分子名称: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase 2, URIDINE-5'-DIPHOSPHATE | | 著者 | Matsumoto, M, Umezawa, A, Kotake, T, Fushinobu, S. | | 登録日 | 2023-10-07 | | 公開日 | 2024-05-08 | | 最終更新日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Cytosolic UDP-L-arabinose synthesis by bifunctional UDP-glucose 4-epimerases in Arabidopsis.
Plant J., 119, 2024
|
|
8UBW
 
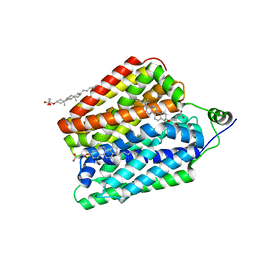 | | Choline-bound FLVCR1 | | 分子名称: | CHOLESTEROL HEMISUCCINATE, CHOLINE ION, Heme transporter FLVCR1 | | 著者 | Hite, R.K, Son, Y. | | 登録日 | 2023-09-25 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.59 Å) | | 主引用文献 | Structural basis of lipid head group entry to the Kennedy pathway by FLVCR1.
Nature, 629, 2024
|
|
8A1A
 
 | | Structure of a leucinostatin derivative determined by host lattice display : L1F11V1 construct | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-(2-methoxyethoxy)-11,15-dimethyl-8-oxa-2,11,15,19,21,23-hexazatetracyclo[15.6.1.13,7.020,24]pentacosa-1(23),3(25),4,6,17,20(24),21-heptaen-10-one, ... | | 著者 | Mittl, P.R.E. | | 登録日 | 2022-06-01 | | 公開日 | 2022-12-07 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structure of a hydrophobic leucinostatin derivative determined by host lattice display.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
