6NMJ
 
 | | Crystal Structure of Rat Ric-8A G alpha binding domain, "Paratone-N Immersed" | | 分子名称: | Resistance to inhibitors of cholinesterase 8 homolog A (C. elegans) | | 著者 | Zeng, B, Mou, T.C, Sprang, S.R. | | 登録日 | 2019-01-11 | | 公開日 | 2019-06-26 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure, Function, and Dynamics of the G alpha Binding Domain of Ric-8A.
Structure, 27, 2019
|
|
6NWS
 
 | | RORgamma Ligand Binding Domain | | 分子名称: | 2-chloro-6-fluoro-N-(1-{[3-(trifluoromethyl)phenyl]sulfonyl}-2,3-dihydro-1H-indol-6-yl)benzamide, Nuclear receptor ROR-gamma | | 著者 | Strutzenberg, T.S, Park, H.J, Griffin, P.R. | | 登録日 | 2019-02-07 | | 公開日 | 2019-07-10 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | HDX-MS reveals structural determinants for ROR gamma hyperactivation by synthetic agonists.
Elife, 8, 2019
|
|
6NWU
 
 | | RORgamma Ligand Binding Domain | | 分子名称: | 6-[(3,5-dichloropyridin-4-yl)methoxy]-1-{[3-(trifluoromethyl)phenyl]sulfonyl}-2,3-dihydro-1H-indole, Nuclear receptor ROR-gamma | | 著者 | Strutzenberg, T.S, Park, H, Griffin, P.R. | | 登録日 | 2019-02-07 | | 公開日 | 2019-07-10 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | HDX-MS reveals structural determinants for ROR gamma hyperactivation by synthetic agonists.
Elife, 8, 2019
|
|
6NWT
 
 | | RORgamma Ligand Binding Domain | | 分子名称: | 1,1,1,3,3,3-hexafluoro-2-[2-fluoro-4'-({4-[(pyridin-4-yl)methyl]piperazin-1-yl}methyl)[1,1'-biphenyl]-4-yl]propan-2-ol, Nuclear receptor ROR-gamma | | 著者 | Strutzenberg, T.S, Park, H, Griffin, P.R. | | 登録日 | 2019-02-07 | | 公開日 | 2019-07-10 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | HDX-MS reveals structural determinants for ROR gamma hyperactivation by synthetic agonists.
Elife, 8, 2019
|
|
4MZ4
 
 | | Discovery of an Irreversible HCV NS5B Polymerase Inhibitor | | 分子名称: | 1-[(2-chloroquinolin-3-yl)methyl]-6-fluoro-5-methyl-3-(2-oxo-1,2-dihydropyridin-3-yl)-1H-indole-2-carboxylic acid, PHOSPHATE ION, RNA-directed RNA polymerase | | 著者 | Zeng, Q, Anilkumar, G.N, Rosenblum, S.B, Huang, H.-C, Lesburg, C.A, Jiang, Y, Selyutin, O, Chan, T.-Y, Bennett, F, Chen, K.X, Venkatraman, S, Sannigrahi, M, Velazquez, F, Duca, J.S, Gavalas, S, Huang, Y, Pu, H, Wang, L, Pinto, P, Vibulbhan, B, Agrawal, S, Ferrari, E, Jiang, C.-K, Li, C, Hesk, D, Gesell, J, Sorota, S, Shih, N.-Y, Njoroge, F.G, Kozlowski, J.A. | | 登録日 | 2013-09-29 | | 公開日 | 2013-12-11 | | 最終更新日 | 2013-12-18 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Discovery of an irreversible HCV NS5B polymerase inhibitor.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
2L5C
 
 | |
6C4H
 
 | |
6CBX
 
 | | Crystal structure of human SET and MYND Domain Containing protein 2 with MTF1497 | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, N-lysine methyltransferase SMYD2, ... | | 著者 | ZENG, H, DONG, A, Hutchinson, A, Seitova, A, TATLOCK, J, KUMPF, R, OWEN, A, TAYLOR, A, Casimiro-Garcia, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | 登録日 | 2018-02-05 | | 公開日 | 2018-03-14 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Crystal structure of human SET and MYND Domain Containing protein 2 with MTF1497
to be published
|
|
5YM8
 
 | |
5YTE
 
 | | Large fragment of DNA Polymerase I from Thermus aquaticus in a closed ternary complex with with natural dT:dATP base pair | | 分子名称: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*TP*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), ... | | 著者 | Zeng, H, Mondal, M, Song, R.Y, Zhang, J, Xia, B, Gao, Y.Q, Yi, C.Q. | | 登録日 | 2017-11-17 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Unnatural Cytosine Bases Recognized as Thymines by DNA Polymerases by the Formation of the Watson-Crick Geometry.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
6CN2
 
 | | Crystal structure of zebrafish Phosphatidylinositol-4-phosphate 5- kinase alpha isoform D236N with bound ATP/Ca2+ | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Phosphatidylinositol-4-phosphate 5-kinase, ... | | 著者 | Zeng, X, Sui, D, Hu, J. | | 登録日 | 2018-03-07 | | 公開日 | 2018-03-21 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.102 Å) | | 主引用文献 | Structural insights into lethal contractural syndrome type 3 (LCCS3) caused by a missense mutation of PIP5K gamma.
Biochem. J., 475, 2018
|
|
5YM6
 
 | |
5Y9H
 
 | | Crystal structure of SafDAA-dsc complex | | 分子名称: | SafD,Putative outer membrane protein,Putative outer membrane protein,Putative outer membrane protein | | 著者 | Zeng, L.H, Zhang, L, Wang, P.R, Meng, G. | | 登録日 | 2017-08-25 | | 公開日 | 2017-11-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural basis of host recognition and biofilm formation by Salmonella Saf pili
Elife, 6, 2017
|
|
5Y9G
 
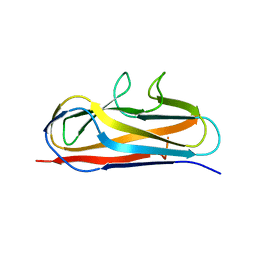 | | Crystal structure of Salmonella SafD adhesin | | 分子名称: | Pilin structural protein SafD,Pilus assembly protein | | 著者 | Zeng, L.H, Zhang, L, Wang, P.R, Meng, G. | | 登録日 | 2017-08-25 | | 公開日 | 2017-11-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural basis of host recognition and biofilm formation by Salmonella Saf pili
Elife, 6, 2017
|
|
5YTD
 
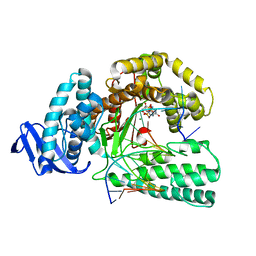 | | large fragment of DNA Polymerase I from Thermus aquaticus in a closed ternary complex with the natural base pair 5fC:dGTP | | 分子名称: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*(5FC)P*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), ... | | 著者 | Zeng, H, Mondal, M, Song, R.Y, Zhang, J, Xia, B, Gao, Y.Q, Yi, C.Q. | | 登録日 | 2017-11-17 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Unnatural Cytosine Bases Recognized as Thymines by DNA Polymerases by the Formation of the Watson-Crick Geometry.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
5YTC
 
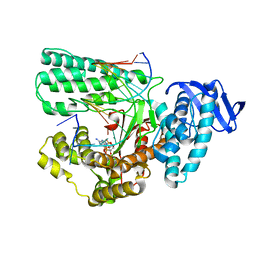 | | Large fragment of DNA Polymerase I from Thermus aquaticus in a closed ternary complex with the unnatural base M-fC pair with dATP in the active site | | 分子名称: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*(92F)P*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC)P*(DOC))-3'), ... | | 著者 | Zeng, H, Mondal, M, Song, R.Y, Zhang, J, Xia, B, Yi, C.Q. | | 登録日 | 2017-11-17 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Unnatural Cytosine Bases Recognized as Thymines by DNA Polymerases by the Formation of the Watson-Crick Geometry.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
5YTF
 
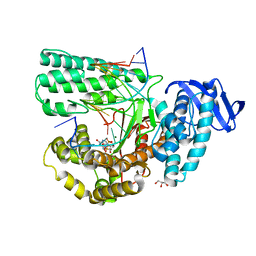 | | Structure of large fragment of DNA Polymerase I from Thermus aquaticus Host-Guest complex with the unnatural base M-fC pair with dA | | 分子名称: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*CP*GP*GP*CP*GP*CP*CP*GP*(92F)P*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), ... | | 著者 | Zeng, H, Mondal, M, Song, R.Y, Zhang, J, Xia, B, Gao, Y.Q, Yi, C.Q. | | 登録日 | 2017-11-17 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Unnatural Cytosine Bases Recognized as Thymines by DNA Polymerases by the Formation of the Watson-Crick Geometry.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
5YTG
 
 | | Structure of large fragment of DNA Polymerase I from Thermus aquaticus Host-Guest complex with the unnatural base I-fC pair with dA | | 分子名称: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*CP*GP*GP*CP*GP*CP*CP*GP*(94O)P*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), ... | | 著者 | Zeng, H, Mondal, M, Song, R.Y, Zhang, J, Xia, B, Gao, Y.Q, Yi, C.Q. | | 登録日 | 2017-11-17 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Unnatural Cytosine Bases Recognized as Thymines by DNA Polymerases by the Formation of the Watson-Crick Geometry.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
5YTH
 
 | | Structure of large fragment of DNA Polymerase I from Thermus aquaticus Host-Guest complex with the unnatural base M-fC pair with dG | | 分子名称: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*CP*GP*GP*CP*GP*CP*CP*GP*(92F)P*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*GP*CP*GP*GP*CP*GP*CP*(DOC))-3'), ... | | 著者 | Zeng, H, Mondal, M, Song, R.Y, Zhang, J, Xia, B, Gao, Y.Q, Yi, C.Q. | | 登録日 | 2017-11-17 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.53 Å) | | 主引用文献 | Unnatural Cytosine Bases Recognized as Thymines by DNA Polymerases by the Formation of the Watson-Crick Geometry.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
3G6Z
 
 | | Design and Preparation of Potent, Non-Peptidic, Bioavailable Renin Inhibitors | | 分子名称: | (1R,5S)-N-cyclopropyl-7-{4-[2-(2,6-dichloro-4-methylphenoxy)ethoxy]phenyl}-N-(2,3-dimethylbenzyl)-3,9-diazabicyclo[3.3.1]non-6-ene-6-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Renin | | 著者 | Bezencon, O, Bur, D, Prade, L, Weller, T, Boss, C, Fischli, W. | | 登録日 | 2009-02-09 | | 公開日 | 2009-06-30 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Design and Preparation of Potent, Nonpeptidic, Bioavailable Renin Inhibitors
J.Med.Chem., 52, 2009
|
|
3G72
 
 | | Design and Preparation of Potent, Non-Peptidic, Bioavailable Renin Inhibitors | | 分子名称: | (1S,5R)-7-{4-[3-(2-chloro-3,6-difluorophenoxy)propyl]phenyl}-N-cyclopropyl-N-(2,3-dichlorobenzyl)-3,9-diazabicyclo[3.3.1]non-6-ene-6-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | 著者 | Bezencon, O, Bur, D, Prade, L, Weller, T, Boss, C, Fischli, W. | | 登録日 | 2009-02-09 | | 公開日 | 2009-06-30 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Design and Preparation of Potent, Nonpeptidic, Bioavailable Renin Inhibitors
J.Med.Chem., 52, 2009
|
|
1D2N
 
 | | D2 DOMAIN OF N-ETHYLMALEIMIDE-SENSITIVE FUSION PROTEIN | | 分子名称: | GLYCEROL, MAGNESIUM ION, N-ETHYLMALEIMIDE-SENSITIVE FUSION PROTEIN, ... | | 著者 | Lenzen, C.U, Steinmann, D, Whiteheart, S.W, Weis, W.I. | | 登録日 | 1998-06-30 | | 公開日 | 1998-10-14 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal structure of the hexamerization domain of N-ethylmaleimide-sensitive fusion protein.
Cell(Cambridge,Mass.), 94, 1998
|
|
3G70
 
 | | Design and Preparation of Potent, Non-Peptidic, Bioavailable Renin Inhibitors | | 分子名称: | (1R,5S)-7-{4-[3-(2-chloro-3,6-difluorophenoxy)propyl]phenyl}-N-cyclopropyl-N-(2,3-dichlorobenzyl)-3,9-diazabicyclo[3.3.1]non-6-ene-6-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | 著者 | Bezencon, O, Bur, D, Prade, L, Weller, T, Boss, C, Fischli, W. | | 登録日 | 2009-02-09 | | 公開日 | 2009-06-30 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Design and Preparation of Potent, Nonpeptidic, Bioavailable Renin Inhibitors
J.Med.Chem., 52, 2009
|
|
6C5L
 
 | |
1W8S
 
 | | The mechanism of the Schiff Base Forming Fructose-1,6-bisphosphate Aldolase: Structural analysis of reaction intermediates | | 分子名称: | 1,6-di-O-phosphono-beta-D-fructofuranose, FRUCTOSE-BISPHOSPHATE ALDOLASE CLASS I | | 著者 | Lorentzen, E, Hensel, R, Siebers, B, Pohl, E. | | 登録日 | 2004-09-27 | | 公開日 | 2005-03-23 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Mechanism of the Schiff base forming fructose-1,6-bisphosphate aldolase: structural analysis of reaction intermediates.
Biochemistry, 44, 2005
|
|
