7GTQ
 
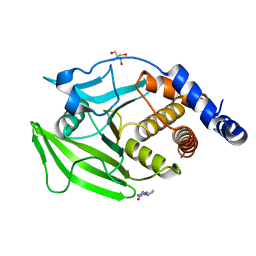 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOPL000311a | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1, ~{N}-(2-ethyl-1,2,3,4-tetrazol-5-yl)butanamide | | 著者 | Mehlman, T, Ginn, H.M, Keedy, D.A. | | 登録日 | 2024-01-03 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
2OFO
 
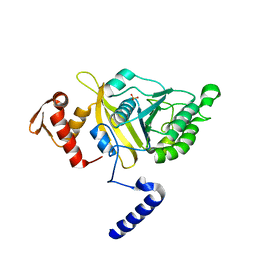 | | MSrecA-native | | 分子名称: | PHOSPHATE ION, Protein recA | | 著者 | Krishna, R, Rajan Prabu, J, Manjunath, G.P, Datta, S, Chandra, N.R, Muniyappa, K, Vijayan, M. | | 登録日 | 2007-01-04 | | 公開日 | 2007-06-19 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.16 Å) | | 主引用文献 | Snapshots of RecA protein involving movement of the C-domain and different conformations of the DNA-binding loops: crystallographic and comparative analysis of 11 structures of Mycobacterium smegmatis RecA
J.Mol.Biol., 367, 2007
|
|
7GSN
 
 | |
7FFE
 
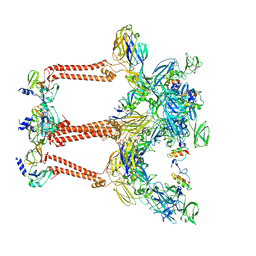 | | Cryo-EM structure of VEEV VLP | | 分子名称: | Capsid protein, Spike glycoprotein E1, Spike glycoprotein E2, ... | | 著者 | Zhang, X, Xiang, Y, Ma, J, Ma, B, Huang, C. | | 登録日 | 2021-07-23 | | 公開日 | 2021-10-20 | | 最終更新日 | 2021-11-17 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structure of Venezuelan equine encephalitis virus with its receptor LDLRAD3.
Nature, 598, 2021
|
|
3IUH
 
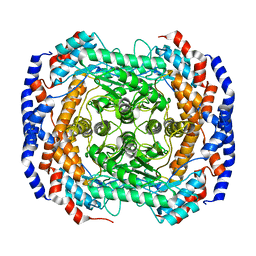 | | Co2+-bound form of Pseudomonas stutzeri L-rhamnose isomerase | | 分子名称: | COBALT (II) ION, L-rhamnose isomerase | | 著者 | Yoshida, H, Yamaji, M, Ishii, T, Izumori, K, Kamitori, S. | | 登録日 | 2009-08-31 | | 公開日 | 2010-02-02 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Catalytic reaction mechanism of Pseudomonas stutzeri l-rhamnose isomerase deduced from X-ray structures
Febs J., 277, 2010
|
|
2OEP
 
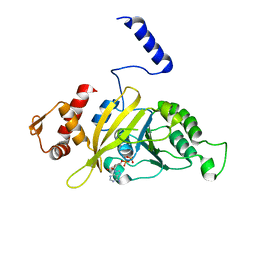 | | MSrecA-ADP-complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Protein recA | | 著者 | Krishna, R, Rajan Prabu, J, Manjunath, G.P, Datta, S, Chandra, N.R, Muniyappa, K, Vijayan, M. | | 登録日 | 2006-12-31 | | 公開日 | 2007-06-19 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Snapshots of RecA protein involving movement of the C-domain and different conformations of the DNA-binding loops: crystallographic and comparative analysis of 11 structures of Mycobacterium smegmatis RecA
J.Mol.Biol., 367, 2007
|
|
7GT2
 
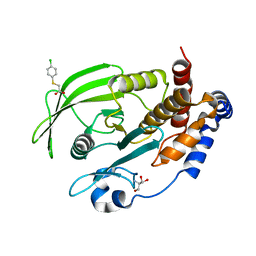 | |
7F3F
 
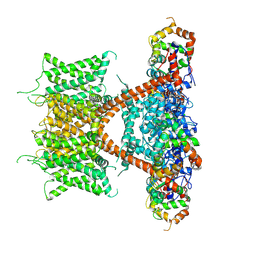 | | CryoEM structure of human Kv4.2-KChIP1 complex | | 分子名称: | Isoform 2 of Kv channel-interacting protein 1, Potassium voltage-gated channel subfamily D member 2 | | 著者 | Kise, Y, Nureki, O. | | 登録日 | 2021-06-16 | | 公開日 | 2021-10-13 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis of gating modulation of Kv4 channel complexes.
Nature, 599, 2021
|
|
7GU7
 
 | |
3BGF
 
 | |
7GTB
 
 | |
7GU9
 
 | |
7GTJ
 
 | |
3IVT
 
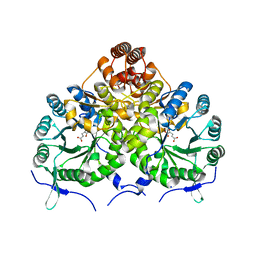 | | Homocitrate Synthase Lys4 bound to 2-OG | | 分子名称: | 2-OXOGLUTARIC ACID, Homocitrate synthase, mitochondrial, ... | | 著者 | Bulfer, S.L, Scott, E.M, Couture, J.-F, Pillus, L, Trievel, R.C. | | 登録日 | 2009-09-01 | | 公開日 | 2009-09-22 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Crystal structure and functional analysis of homocitrate synthase, an essential enzyme in lysine biosynthesis.
J.Biol.Chem., 284, 2009
|
|
7GTX
 
 | |
7F2E
 
 | |
3BH7
 
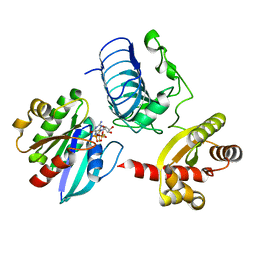 | | Crystal structure of the RP2-Arl3 complex bound to GDP-AlF4 | | 分子名称: | ADP-ribosylation factor-like protein 3, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Veltel, S, Gasper, R, Wittinghofer, A. | | 登録日 | 2007-11-28 | | 公開日 | 2008-03-25 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The retinitis pigmentosa 2 gene product is a GTPase-activating protein for Arf-like 3
Nat.Struct.Mol.Biol., 15, 2008
|
|
7GTV
 
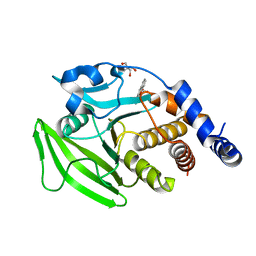 | |
7FFL
 
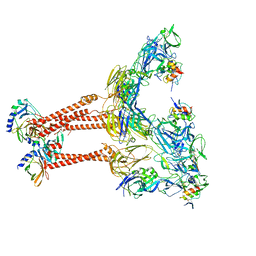 | | Cryo-EM structure of VEEV VLP-LDLRAD3-D1 complex at the 2-fold axes | | 分子名称: | CALCIUM ION, Capsid protein, Low-density lipoprotein receptor class A domain-containing protein 3, ... | | 著者 | Zhang, X, Xiang, Y, Ma, J, Ma, B, Huang, C. | | 登録日 | 2021-07-23 | | 公開日 | 2021-10-20 | | 最終更新日 | 2021-11-17 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structure of Venezuelan equine encephalitis virus with its receptor LDLRAD3.
Nature, 598, 2021
|
|
7MTX
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Bacillus anthracis in the complex with IMP and the inhibitor P176 | | 分子名称: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, N-{2-chloro-5-[({2-[3-(prop-1-en-2-yl)phenyl]propan-2-yl}carbamoyl)amino]phenyl}-beta-D-ribopyranosylamine, ... | | 著者 | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Gu, M, Gollapalli, D, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-05-13 | | 公開日 | 2021-06-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from
Bacillus anthracis in the complex with IMP and the inhibitor P176
To Be Published
|
|
7FFF
 
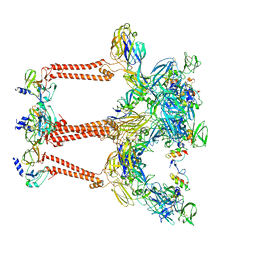 | | Structure of Venezuelan equine encephalitis virus with the receptor LDLRAD3 | | 分子名称: | CALCIUM ION, Capsid protein, Low-density lipoprotein receptor class A domain-containing protein 3, ... | | 著者 | Zhang, X, Xiang, Y, Ma, J, Ma, B, Huang, C. | | 登録日 | 2021-07-23 | | 公開日 | 2021-10-20 | | 最終更新日 | 2021-11-17 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structure of Venezuelan equine encephalitis virus with its receptor LDLRAD3.
Nature, 598, 2021
|
|
7MTU
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Bacillus anthracis in the complex with IMP and the inhibitor P221 | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, INOSINIC ACID, ... | | 著者 | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Gu, M, Gollapalli, D, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-05-13 | | 公開日 | 2021-06-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from
Bacillus anthracis in the complex with IMP and the inhibitor P221
To Be Published
|
|
7EP9
 
 | |
3IZH
 
 | | Mm-cpn D386A with ATP | | 分子名称: | Chaperonin | | 著者 | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | 登録日 | 2010-10-29 | | 公開日 | 2011-02-02 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (11 Å) | | 主引用文献 | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
2OGZ
 
 | |
