8DYX
 
 | |
7YQ3
 
 | | human insulin receptor bound with A43 DNA aptamer and insulin | | 分子名称: | IR-A43 aptamer, Insulin A chain, Insulin, ... | | 著者 | Kim, J, Yunn, N, Ryu, S, Cho, Y. | | 登録日 | 2022-08-05 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Functional selectivity of insulin receptor revealed by aptamer-trapped receptor structures.
Nat Commun, 13, 2022
|
|
7YQ4
 
 | | human insulin receptor bound with A62 DNA aptamer and insulin - locally refined | | 分子名称: | IR-A62 aptamer, Insulin A chain, Insulin, ... | | 著者 | Kim, J, Yunn, N, Ryu, S, Cho, Y. | | 登録日 | 2022-08-05 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.95 Å) | | 主引用文献 | Functional selectivity of insulin receptor revealed by aptamer-trapped receptor structures.
Nat Commun, 13, 2022
|
|
7YQ6
 
 | | human insulin receptor bound with A62 DNA aptamer | | 分子名称: | IR-A62 aptamer, Isoform Short of Insulin receptor | | 著者 | Kim, J, Yunn, N, Ryu, S, Cho, Y. | | 登録日 | 2022-08-05 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.18 Å) | | 主引用文献 | Functional selectivity of insulin receptor revealed by aptamer-trapped receptor structures.
Nat Commun, 13, 2022
|
|
7YQ5
 
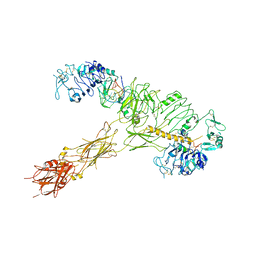 | | human insulin receptor bound with A62 DNA aptamer and insulin | | 分子名称: | IR-A62 aptamer, Insulin A chain, Insulin, ... | | 著者 | Kim, J, Yunn, N, Ryu, S, Cho, Y. | | 登録日 | 2022-08-05 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.27 Å) | | 主引用文献 | Functional selectivity of insulin receptor revealed by aptamer-trapped receptor structures.
Nat Commun, 13, 2022
|
|
7YPZ
 
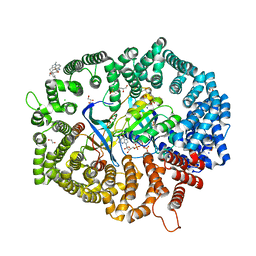 | | Zafirlukast in complex with CRM1-Ran-RanBP1 | | 分子名称: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, ... | | 著者 | Sun, Q, Lei, Y. | | 登録日 | 2022-08-05 | | 公開日 | 2023-08-16 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Searching for Novel Noncovalent Nuclear Export Inhibitors through a Drug Repurposing Approach.
J.Med.Chem., 66, 2023
|
|
8DYZ
 
 | |
7YPP
 
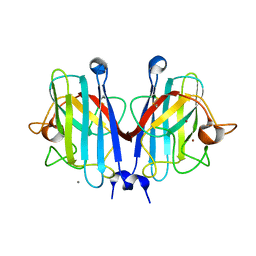 | | Structural basis of a superoxide dismutase from a tardigrade, Ramazzottius varieornatus strain YOKOZUNA-1. | | 分子名称: | CALCIUM ION, COPPER (II) ION, Superoxide dismutase [Cu-Zn], ... | | 著者 | Sim, K.-S, Fukuda, Y, Inoue, T. | | 登録日 | 2022-08-04 | | 公開日 | 2023-07-26 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of a superoxide dismutase from a tardigrade: Ramazzottius varieornatus strain YOKOZUNA-1.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
7YPR
 
 | | Structural basis of a superoxide dismutase from a tardigrade, Ramazzottius varieornatus strain YOKOZUNA-1. | | 分子名称: | COPPER (II) ION, POTASSIUM ION, Superoxide dismutase [Cu-Zn], ... | | 著者 | Sim, K.-S, Fukuda, Y, Inoue, T. | | 登録日 | 2022-08-04 | | 公開日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.101 Å) | | 主引用文献 | Structure of a superoxide dismutase from a tardigrade: Ramazzottius varieornatus strain YOKOZUNA-1.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
8AMZ
 
 | | Spinach 19S proteasome | | 分子名称: | 26S proteasome non-ATPase regulatory subunit 1 homolog, 26S proteasome non-ATPase regulatory subunit 2 homolog, 26S proteasome regulatory subunit 7, ... | | 著者 | Kandolf, S, Grishkovskaya, I, Meinhart, A, Haselbach, D. | | 登録日 | 2022-08-04 | | 公開日 | 2022-08-24 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Cryo-EM structure of the plant 26S proteasome
Plant Communications, 3, 2022
|
|
8DYO
 
 | | Cryo-EM structure of Importin-4 bound to RanGTP | | 分子名称: | GTP-binding nuclear protein GSP1/CNR1, GUANOSINE-5'-TRIPHOSPHATE, Importin-4, ... | | 著者 | Bernardes, N.E, Fung, H.Y.J, Li, Y, Chen, Z, Chook, Y.M. | | 登録日 | 2022-08-04 | | 公開日 | 2022-09-21 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (7.1 Å) | | 主引用文献 | Structure of IMPORTIN-4 bound to the H3-H4-ASF1 histone-histone chaperone complex.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DYC
 
 | | Human CYP3A4 bound to a substrate | | 分子名称: | 2-butyl-6-(butylamino)-1H-benzo[de]isoquinoline-1,3(2H)-dione, Cytochrome P450 3A4, GLYCEROL, ... | | 著者 | Sevrioukova, I.F. | | 登録日 | 2022-08-04 | | 公開日 | 2022-10-26 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal Structure of CYP3A4 Complexed with Fluorol Identifies the Substrate Access Channel as a High-Affinity Ligand Binding Site.
Int J Mol Sci, 23, 2022
|
|
8AMR
 
 | | Coiled-coil domain of human TRIM3 | | 分子名称: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, GLYCEROL, Tripartite motif-containing protein 3 | | 著者 | Perez-Borrajero, C, Murciano, B, Hennig, J. | | 登録日 | 2022-08-04 | | 公開日 | 2023-11-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Structural and biophysical studies of TRIM2 and TRIM3
To Be Published
|
|
8DYB
 
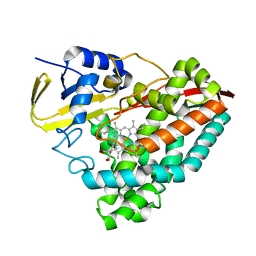 | |
7YPJ
 
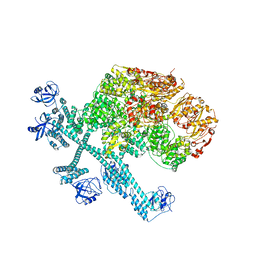 | | Spiral pentamer of the substrate-free Lon protease with a S678A mutation | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Lon protease | | 著者 | Li, S, Hsieh, K.Y, Kuo, C.I, Lee, S.H, Ho, M.R, Wang, C.H, Zhang, K, Chang, C.I. | | 登録日 | 2022-08-03 | | 公開日 | 2023-10-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YPK
 
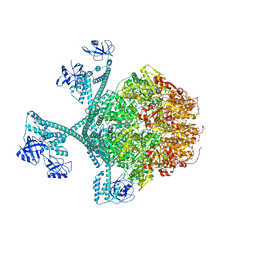 | | Close-ring hexamer of the substrate-bound Lon protease with an S678A mutation | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Lon protease, alpha-S1-casein | | 著者 | Li, S, Hsieh, K.Y, Kuo, C.I, Lee, S.H, Ho, M.R, Wang, C.H, Zhang, K, Chang, C.I. | | 登録日 | 2022-08-03 | | 公開日 | 2023-10-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YPI
 
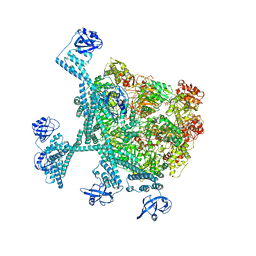 | | Spiral hexamer of the substrate-free Lon protease with a Y224S mutation | | 分子名称: | Lon protease, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | 著者 | Li, S, Hsieh, K.Y, Kuo, C.I, Lee, S.H, Ho, M.R, Wang, C.H, Zhang, K, Chang, C.I. | | 登録日 | 2022-08-03 | | 公開日 | 2023-10-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YPH
 
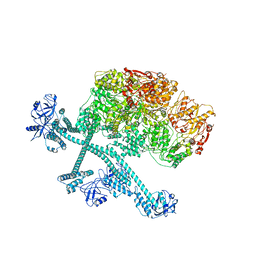 | | Open-spiral pentamer of the substrate-free Lon protease with a Y224S mutation | | 分子名称: | Lon protease, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | 著者 | Li, S, Hsieh, K.Y, Kuo, C.I, Lee, S.H, Ho, M.R, Wang, C.H, Zhang, K, Chang, C.I. | | 登録日 | 2022-08-03 | | 公開日 | 2023-10-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | ELECTRON MICROSCOPY (3.68 Å) | | 主引用文献 | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
8AMO
 
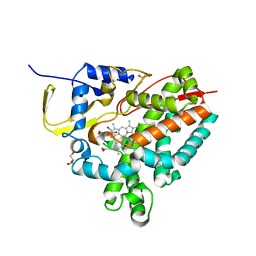 | | Crystal structure of M. tuberculosis CYP143 | | 分子名称: | CHLORIDE ION, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Bukhdruker, S, Varaksa, T, Grudo, A, Marin, E, Kapranov, I, Shevtsov, M, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | 登録日 | 2022-08-03 | | 公開日 | 2023-02-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural insights into 3Fe-4S ferredoxins diversity in M. tuberculosis highlighted by a first redox complex with P450.
Front Mol Biosci, 9, 2022
|
|
8AMQ
 
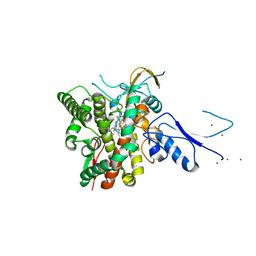 | | Crystal structure of the complex CYP143-FdxE from M. tuberculosis | | 分子名称: | FE3-S4 CLUSTER, NICKEL (II) ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Bukhdruker, S, Varaksa, T, Smolskaya, S, Marin, E, Kapranov, I, Kovalev, K, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | 登録日 | 2022-08-03 | | 公開日 | 2023-02-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural insights into 3Fe-4S ferredoxins diversity in M. tuberculosis highlighted by a first redox complex with P450.
Front Mol Biosci, 9, 2022
|
|
8AMC
 
 | | Crystal Structure of cat allergen Fel d 4 | | 分子名称: | Allergen Fel d 4, SULFATE ION | | 著者 | Schooltink, L, Sagmeister, T, Gottstein, N, Pavkov-Keller, T, Keller, W. | | 登録日 | 2022-08-03 | | 公開日 | 2023-08-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Crystal Structure of cat allergen Fel d 4
To Be Published
|
|
8DXB
 
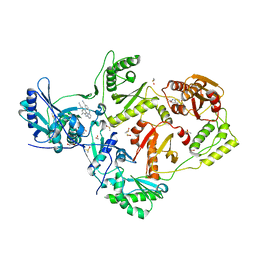 | | HIV-1 reverse transcriptase/rilpivirine with bound fragment 5-fluoroindole-2-carboxylic acid at the NNRTI Adjacent site | | 分子名称: | 1,2-ETHANEDIOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, 5-fluoroindole-2-carboxylic acid, ... | | 著者 | Chopra, A, Ruiz, F.X, Bauman, J.D, Arnold, E. | | 登録日 | 2022-08-02 | | 公開日 | 2023-05-31 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Halo Library, a Tool for Rapid Identification of Ligand Binding Sites on Proteins Using Crystallographic Fragment Screening.
J.Med.Chem., 66, 2023
|
|
8DXH
 
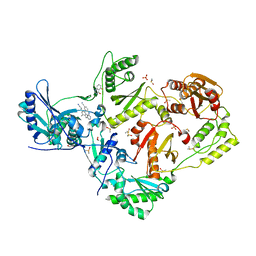 | | HIV-1 reverse transcriptase/rilpivirine with bound fragment 5-fluoroquinazolin-4-ol at W266 site | | 分子名称: | 1,2-ETHANEDIOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, 5-fluoranyl-3,4-dihydroquinazolin-4-ol, ... | | 著者 | Chopra, A, Ruiz, F.X, Bauman, J.D, Arnold, E. | | 登録日 | 2022-08-02 | | 公開日 | 2023-05-31 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Halo Library, a Tool for Rapid Identification of Ligand Binding Sites on Proteins Using Crystallographic Fragment Screening.
J.Med.Chem., 66, 2023
|
|
8DX8
 
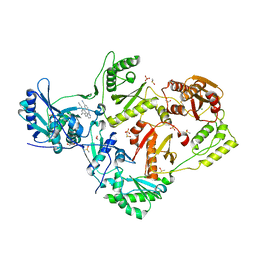 | | HIV-1 reverse transcriptase/rilpivirine with bound fragment 2-chloro-6-fluorophenethylamine at the 415 site | | 分子名称: | 1,2-ETHANEDIOL, 2-(2-chloro-6-fluorophenyl)ethan-1-amine, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, ... | | 著者 | Chopra, A, Ruiz, F.X, Bauman, J.D, Arnold, E. | | 登録日 | 2022-08-02 | | 公開日 | 2023-05-31 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Halo Library, a Tool for Rapid Identification of Ligand Binding Sites on Proteins Using Crystallographic Fragment Screening.
J.Med.Chem., 66, 2023
|
|
8DXM
 
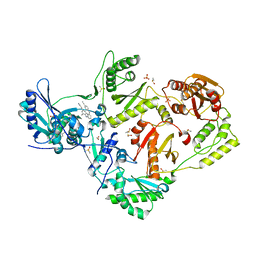 | | HIV-1 reverse transcriptase/rilpivirine with bound fragment 4-bromophenol at the Knuckles site | | 分子名称: | 1,2-ETHANEDIOL, 4-BROMOPHENOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, ... | | 著者 | Chopra, A, Ruiz, F.X, Bauman, J.D, Arnold, E. | | 登録日 | 2022-08-02 | | 公開日 | 2023-05-31 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Halo Library, a Tool for Rapid Identification of Ligand Binding Sites on Proteins Using Crystallographic Fragment Screening.
J.Med.Chem., 66, 2023
|
|
